This study has used a combination of clinical information, spoligotyping, and georeferencing system to elucidate the genetic diversity of the Mycobacterium tuberculosis isolates circulating in a TB-prevalent municipality of Northeast Brazil.
MethodsA total of 115 M. tuberculosis strains were isolated from pulmonary tuberculosis patients from January 2007 to March 2008 in Fortaleza. Drug susceptibility and spoligotyping assays were performed and place of residence of the patients were georeferenced.
ResultsOf the M. tuberculosis strains studied, 51 (44.3%) isolates were resistant to at least one drug (R-TB) and 64 (55.7%) were sensitive to all the drugs tested (S-TB). A high frequency of resistance was found in previously treated cases (84%) and among new cases (16%; p<0.001). A total of 74 (64%) isolates were grouped into 22 spoligotyped lineages, while 41 (36%) isolates were identified as new. Among the predominant genotypes, 33% were Latim American Mediterranean (LAM), 12% Haarlem (H), and 5% U. There was no association of geographic distribution of RT-TB patients as compared to the controls and also the geographic location to the spoligotype patterns. The geospatial analysis revealed that 24 (23%) patients (hot spot zones) either shared the same residence or lived in a close neighborhood of a case. Among these concentration zones, the patients lived in the same residence and shared a common genotype pattern and resistance pattern.
DiscussionIt was observed that the spoligopatterns family distribution was similar to that reported for South America, prevailing the LAM and H lineages. A high rate-case among the resistant TB group occurs as a result of transmitted and acquired resistance. A more effective surveillance program is needed in order to succeed in reducing tuberculosis in Northeast Brazil.
Drug-resistant Mycobacterium tuberculosis (R-TB) is widespread in the world, and of particular concern is the occurrence of multidrug-resistant tuberculosis (MDR-TB), defined as resistance to at least isoniazid (INH) and rifampicin (RIF).1 Resistance may result either from a selection of drug-resistant organisms (acquired resistance) or infection with a drug-resistant TB strain (primary resistance). Acquired resistance is responsible for the majority of the cases and is well-associated to prior-exposure to anti-TB drugs, which includes those who have relapsed, failed and abandoned treatment.2
Among all incident cases of TB globally, 3.6% are estimated to have MDR-TB and 27 countries account for 85% of the MDR-TB cases.3 Despite continuing to be one of the high TB burden countries, the prevalence of MDR-TB in Brazil has been estimated to be low;4 surveillance for drug resistance has been limited.5 In 2009, approximately 87,000 TB cases were reported in Brazil with an incidence rate of 45/100,000,4 while in the state of Ceará, in 2010, 3430 new cases were reported with an incidence of 40.1/100,000.6 Of these TB patients, 87.2% were pulmonary TB. Fortaleza is the capital city of Ceará and is the fifth largest city in Brazil, with 2.5 million inhabitants. In Fortaleza, the TB incidence has shown a decrease of 1916 in 2005 to 1651 in 2010.6 Thus, implementation and intensification of new strategies in the control program is required in this region.
Rapid diagnosis, adequate treatment, and contact tracing to arrest further transmission are important factors in the control of tuberculosis. Molecular genotyping can be applied at the population level, and the obtained clustering of isolates provides clues about the patterns and dynamics of transmission in the population. Moreover when genotyping is coupled with geographic information system (GIS), the generated data can assess the spatial epidemiology of the disease.7,8 The geographical information can be correlated with other important data, including clinical, drug resistance and genotyping, assisting localization of diseases and graphical analysis of epidemiological indicators.
There are limited available data regarding drug resistance, transmission pattern and strain distribution in Northeast Brazil. Previous studies of TB in Brazil have been mostly based on isolated information, including drug resistance, identifying risk factors or genotyping patterns.9–11 A combined study based on clinical data, drug resistance patterns, genotyping and GIS has not been previously determined. Thus, we have conducted a study to elucidate the genetic diversity of the M. tuberculosis isolates circulating in a TB-prevalent municipality of Northeast Brazil.
MethodsStudy populationThis was a case–control prospective study conducted from January 2007 to March 2008 in Fortaleza, Northeast Brazil. Participants were recruited at Hospital of Messejana, which is a reference hospital for TB treatment. On an average, Hospital of Messejana cares for 130 TB patients per month. The patients were eligible to participate in the study if they were at least 15 years of age, and had respiratory symptoms or chest X-ray results suggesting clinical pulmonary TB.12 HIV-infected patients, pregnant woman and those with no place of residence were excluded. M. tuberculosis was confirmed by culture from all the eligible participants. One hundred fifteen participants entered the study. A reduced number of participants as compared to the expected were due to the outpatient clinic of the research physician being open for only one day a week. No evidence of M. tuberculosis growth or culture contamination also reduced the total number of participants. Patients with drug resistant strain to at least one anti-TB drug (R-TB), were considered as cases. The control group consisted of all remaining patients infected by fully sensitive M. tuberculosis (S-TB).
A standardized form was used to collect data regarding age, gender, marital status, social behavior and economic status, clinical, radiological and laboratory tests. All these listed factors, and others not listed, had been previously identified to be related to TB.
Ethical considerationsThis project was approved by the Ethical Committee of the Universidade Federal do Ceará, Fortaleza, CE, Brazil. Written consent was obtained from each participant.
M. tuberculosis isolation and drug sensitivity testingSputum specimens were examined by microscopy and cultured, after decontamination with NaOH and inoculation in Lowestein–Jensen medium. All the isolates were identified by microbiological methods.13 Susceptibility testing for isoniazid (INH) (0.2μg/mL), rifampicin (RMP) (40μg/mL), streptomycin (SM) (4μg/mL) and ethambutol (EMB) (2μg/mL) was performed by using the standard proportion method.14
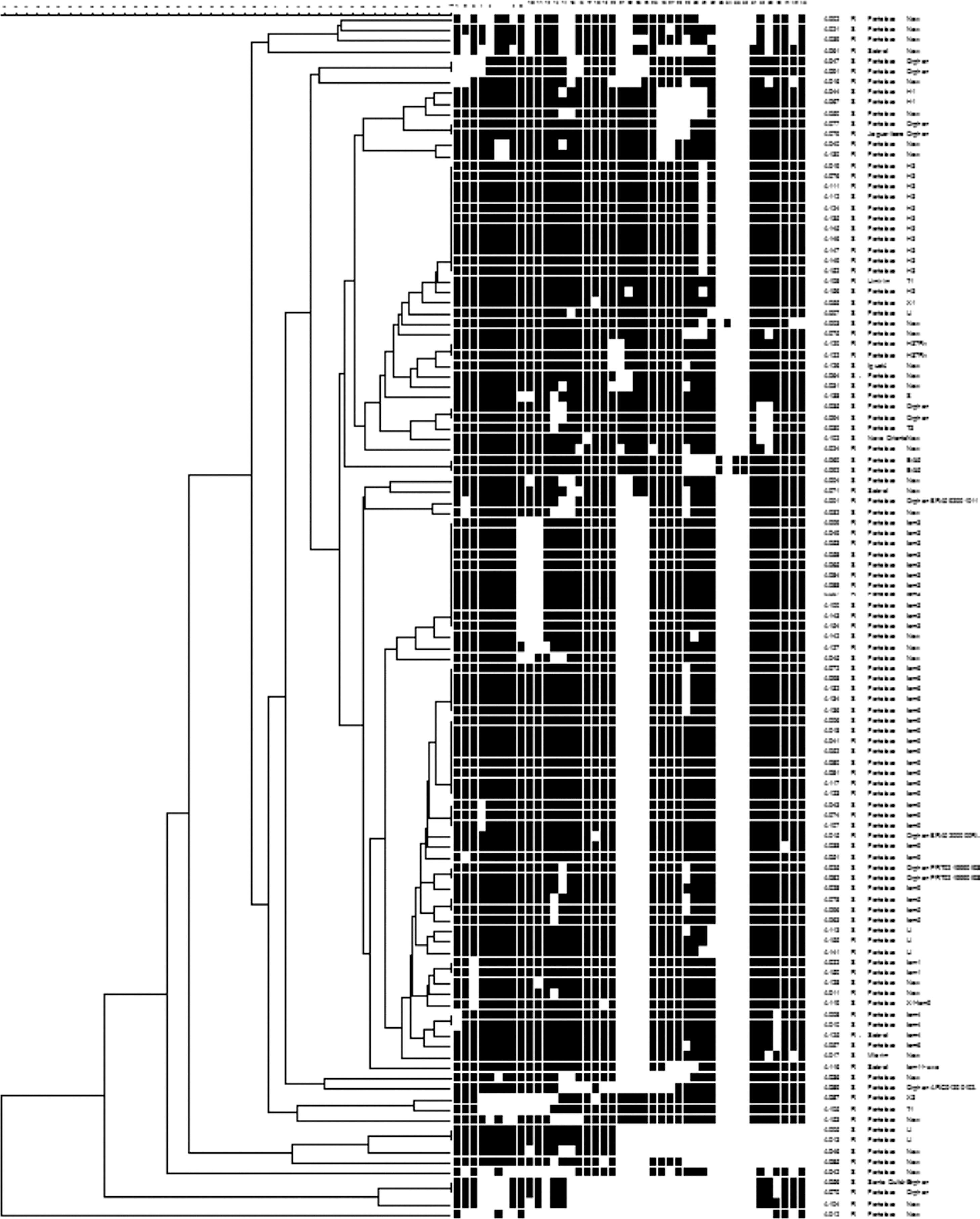
DNA extraction and spoligotypingDNA extraction was performed by using the hexadecyltrimethylammonium bromide (CTAB) method,15,16 and isolates were subjected to spoligotyping.17 The spoligotypes patterns were converted into binary and octal formats according to Brudey et al.18 Spoligopatterns were further analyzed by comparing with SpolDB4 that has been created and maintained at the Institute Pasteur of Guadeloupe.18,19 Spoligotyping results were entered in the Bionumerics Software. Dendograms were generated using the unweighted pairgroup method with arithmetic averages (UPGMA) calculation. The strains with spoligotype similar to any pattern of an M. tuberculosis strain already found in the database were labeled with a defined shared identification, while a spoligotyping exhibiting a profile not yet found was termed “new”.
Geographic information systemPatients were asked to provide their residential addresses and reference points. Two research assistants were blinded to the names of the participants and each patient residence was geo-referenced twice using a GPS handheld device (eTrex®, Garmin).
The coordinates gathered in the field were transcribed to the Excel spreadsheet and subsequently transformed into PDF files to be used directly in the GIS application ArcGIS 9.3 software. After the data had been formatted in spreadsheets, the ArcMap application was used to make the information available in maps showing the distribution of the georeferenced cases in Fortaleza according to drug resistance and identified family lineage.
Statistical and data analysisData were entered in a spreadsheet using Microsoft Excel (2007) and transferred to SSPS 10.0 version and Epi-info 6.04d for analysis. The Student t-test was used to compare the means of continuous variables. A non-parametric test (Mann–Whitney test) was also used to compare data with high asymmetry. Univariate analyses of the association of clinical, laboratory and socio-demographic categorical variables were performed using the Chi-square and Fisher tests. The level of significance was p<0.05.
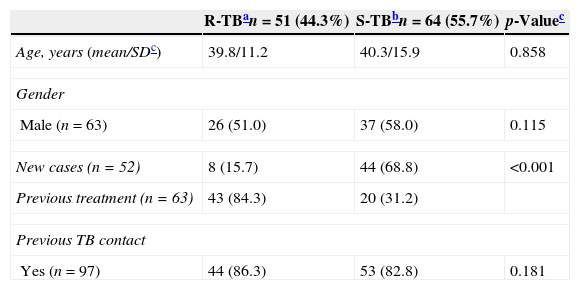
ResultsA total of one hundred fifteen M. tuberculosis clinical isolates from pulmonary sources were studied, with 63 (54.8%) males and 52 (45.2%) females (Table 1). Of these, 51 (44.3%) isolates were R-TB and 64 (55.7%) were S-TB. The average age of the R-TB was 39.8 years (SD 11.2 years; range 20–62 years), while it was 40.3 years (SD 15.9 years; range 19–82 years) among the S-TB. Although resistance was not associated to gender, previously treated cases were more likely among resistant patients (84.3%), when compared to the control group (31.2%, p<0.001) (Table 1). Both R-TB and S-TB groups have had history of previous contact with a TB case, with no significant difference between the two groups (p=0.181).
Demographic profile and other characteristics of 115 patients with pulmonary tuberculosis.
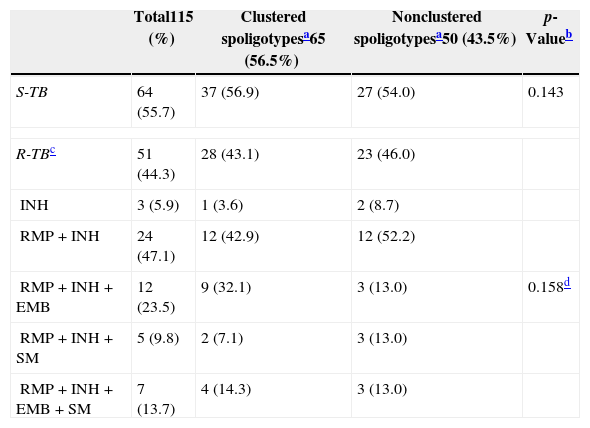
Overall, 65 (56.5%) clusters of spoligopatterns were shared by two or more patients (65 clusters containing 2–13 isolates). Other 50 (43.5%) isolates had unique patterns, designated as nonclustered (Table 2). No significant association was seen among clustered spoligotypes and non-clustered spoligotypes and resistance to anti-TB drugs (p=0.143). Of the 51 R-TB cases, 3 (5.9%) were monoresistant to INH, while the remaining 48 (94.1%) were TB-MDR, being more frequent the resistance to RMP and INH (24/51, 47.1%). None of the isolates had shown resistance to PZA.
Resistance pattern to first line drugs and spoligopatterns among M. tuberculosis isolates from Fortaleza, Brazil.
| Total115 (%) | Clustered spoligotypesa65 (56.5%) | Nonclustered spoligotypesa50 (43.5%) | p-Valueb | |
|---|---|---|---|---|
| S-TB | 64 (55.7) | 37 (56.9) | 27 (54.0) | 0.143 |
| R-TBc | 51 (44.3) | 28 (43.1) | 23 (46.0) | |
| INH | 3 (5.9) | 1 (3.6) | 2 (8.7) | |
| RMP+INH | 24 (47.1) | 12 (42.9) | 12 (52.2) | |
| RMP+INH+EMB | 12 (23.5) | 9 (32.1) | 3 (13.0) | 0.158d |
| RMP+INH+SM | 5 (9.8) | 2 (7.1) | 3 (13.0) | |
| RMP+INH+EMB+SM | 7 (13.7) | 4 (14.3) | 3 (13.0) | |
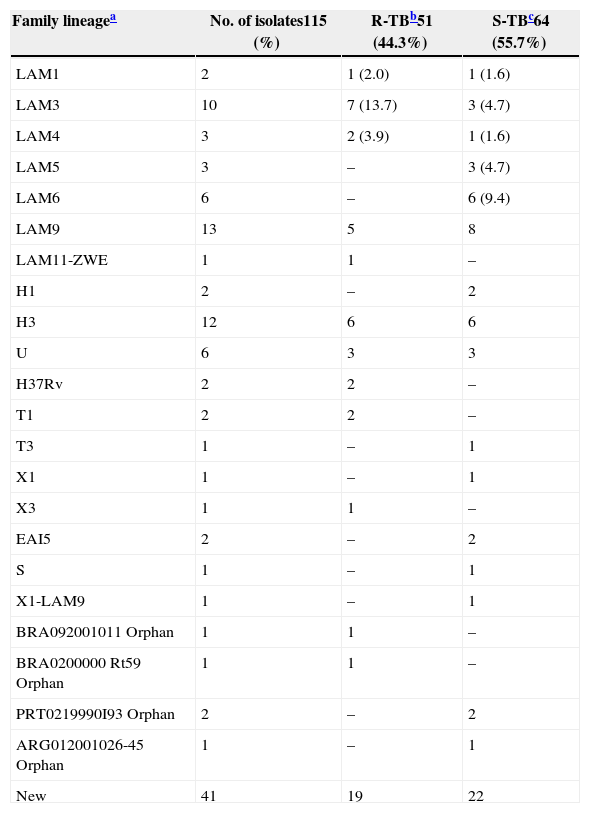
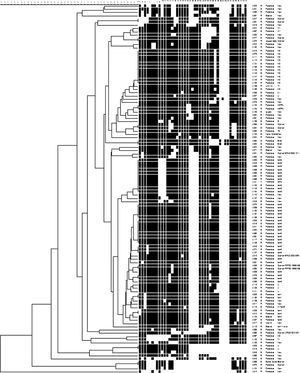
The 115 M. tuberculosis strains were spoligotyped and their data analyzed using UPGMA calculation in the Bionumerics software (Table 3 and Fig. 1). The most frequent families found in the study were Latin American Mediterranean (LAM) (38, 33.0%), Haarlem (H) (14, 12.2%), and U (6, 5.2%). The LAM family was represented by: 13 (34.2%) LAM9, 10 (26.3%) LAM3, 6 (15.8%) LAM6, 3 (7.9%) LAM4, 3 (7.9%) LAM5, 2 (5.3%) LAM1, and 1 (2.6%) LAM11-ZWE. The LAM3 family was in the higher frequency (7 isolates, 43.8%) in the R-TB group as compared to the controls (3, 13.6%), while LAM9 (8 isolates, 36.4%) was more common in the control group compared to the R-TB (5, 31.3%). There were no LAM5 and LAM6 isolates among the R-TB cases. The H family comprised 12 (10.5%) H3 and 2 (1.7%) H1. Families East-African-Indian (EAI), H37Rv, Sicily-Sardinia (S), Tuscany (T), and X comprised 2 (1.7%), 2 (1.7%), 1 (0.9%), 3 (2.6%), and 2 (1.7%) strains, respectively.
Distribution of spoligotyping patterns of 115 tuberculosis cases.
| Family lineagea | No. of isolates115 (%) | R-TBb51 (44.3%) | S-TBc64 (55.7%) |
|---|---|---|---|
| LAM1 | 2 | 1 (2.0) | 1 (1.6) |
| LAM3 | 10 | 7 (13.7) | 3 (4.7) |
| LAM4 | 3 | 2 (3.9) | 1 (1.6) |
| LAM5 | 3 | – | 3 (4.7) |
| LAM6 | 6 | – | 6 (9.4) |
| LAM9 | 13 | 5 | 8 |
| LAM11-ZWE | 1 | 1 | – |
| H1 | 2 | – | 2 |
| H3 | 12 | 6 | 6 |
| U | 6 | 3 | 3 |
| H37Rv | 2 | 2 | – |
| T1 | 2 | 2 | – |
| T3 | 1 | – | 1 |
| X1 | 1 | – | 1 |
| X3 | 1 | 1 | – |
| EAI5 | 2 | – | 2 |
| S | 1 | – | 1 |
| X1-LAM9 | 1 | – | 1 |
| BRA092001011 Orphan | 1 | 1 | – |
| BRA0200000 Rt59 Orphan | 1 | 1 | – |
| PRT0219990I93 Orphan | 2 | – | 2 |
| ARG012001026-45 Orphan | 1 | – | 1 |
| New | 41 | 19 | 22 |
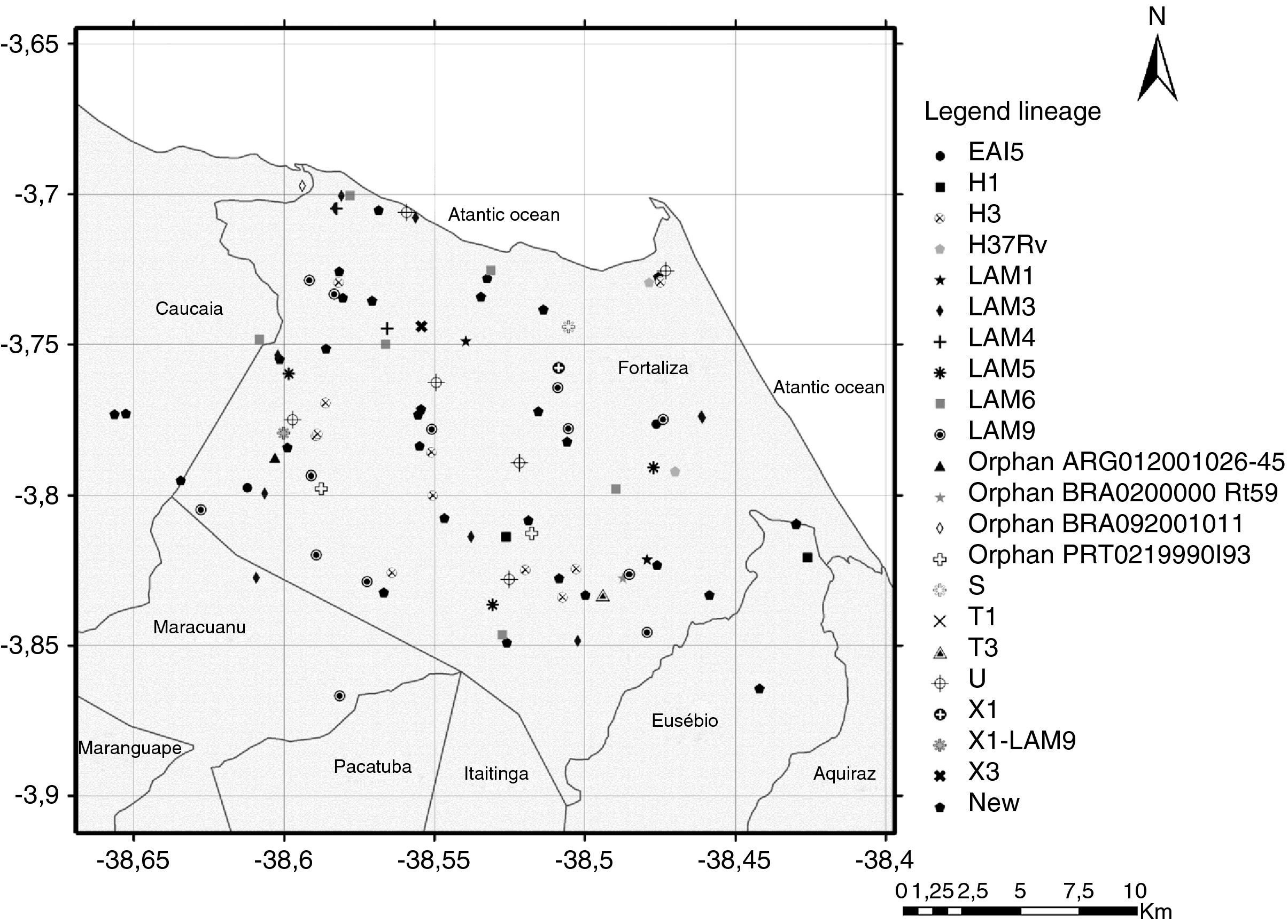
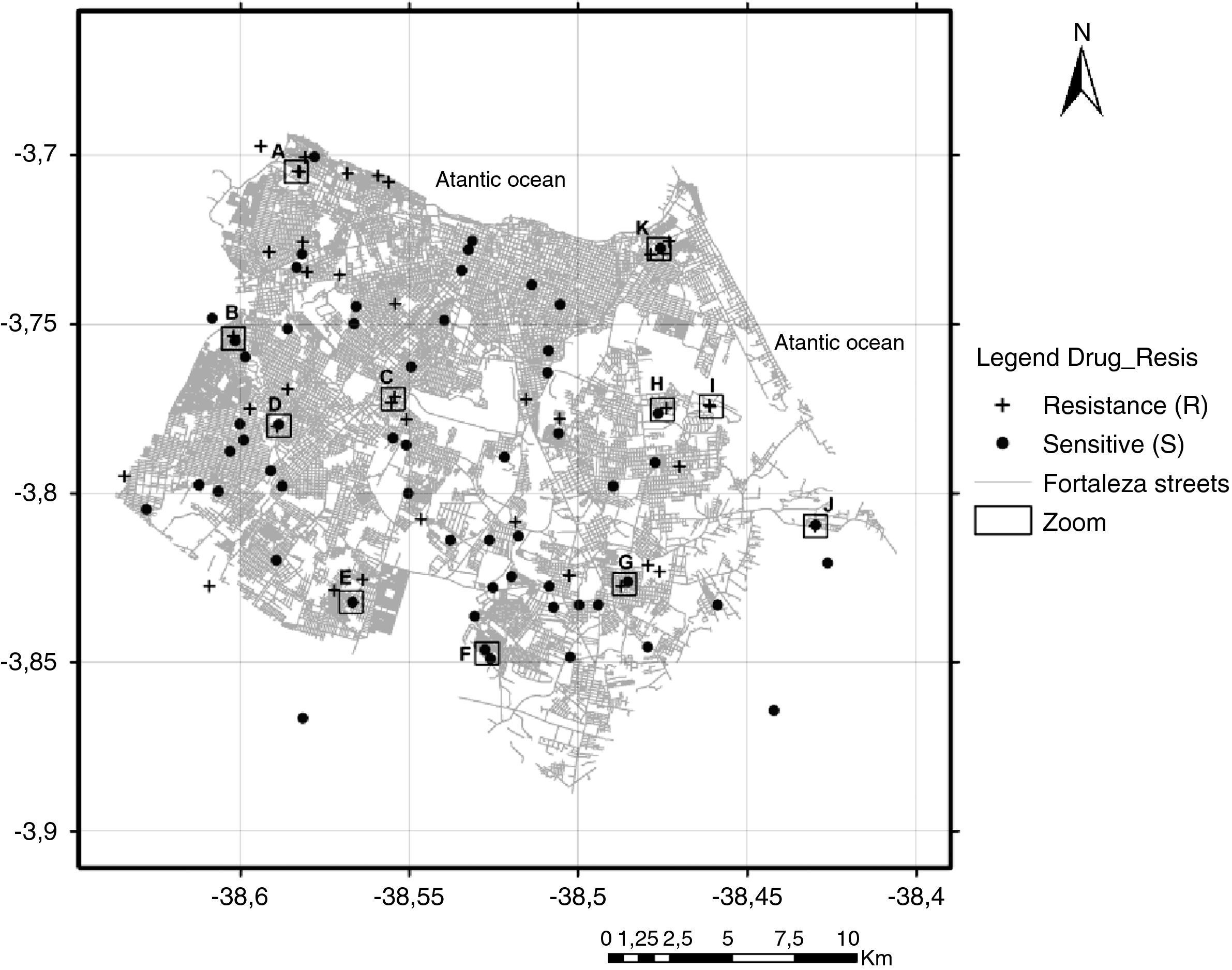
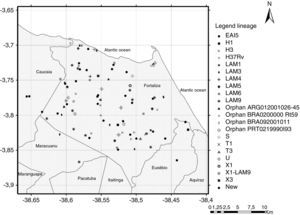
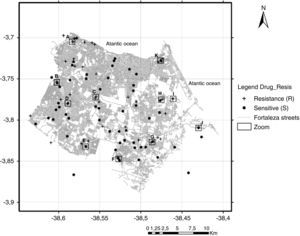
Although the reference hospital was in Fortaleza, 10 (8.7%) patients did not live within the municipality, including two cases that lived in the states of Maranhão and Piauí, resulting in a final geographic location of 105 pulmonary TB isolates. Two maps illustrate reported household locations related to the family lineage identification (Fig. 2) and to the resistance to anti-TB drugs (Fig. 3). The majority of the cases lived in peripheral districts or in poor quality residences of Fortaleza city. The analysis did not indicate differences among the geographic distribution of RT-TB group as compared to controls. In the same way, there was no concentration of spoligotype patterns and geographic location.
Map of the municipality of Fortaleza showing the geographic origin of 105 M. tuberculosis isolates included in the study, 2007–2008. District boundaries are marked with black thin outline. The identified family lineages are represented in the legend (strain genotypes as listed in Fig. 1).
Map of the municipality of Fortaleza showing the 105 M. tuberculosis isolates included in the study by residential location, 2007–2008. +, resistant TB isolates; (●) sensitive TB isolates; (□) squares with letters (A–K) represent TB patients located either in the same residence or in a close neighborhood.
A hot spot analysis of the patients mapped by their residential locations revealed that 24 (22.9%) patients either shared the same residence or lived in a close neighborhood, less than 100 meters of distance (Figs. 2 and 3). These overlapped patients’ zones were labeled by alphabetical letters (A–K), corresponding to 11 spots (Fig. 3). Among these 11 zones, the patients of spots A, D, E and I lived in the same residence and shared a common genotype pattern (A and I, LAM3; D, H3 and E, a new cluster). In addition, the spots A and I also shared the same multidrug-resistance pattern (A, RMP+INH+EMB; I, RMP+INH+SM), whereas the other two spots (D and E), were from S-TB and R-TB patients.
DiscussionIn this study, the genotypic diversity and drug resistance of M. tuberculosis strains in Northeast Brazil, and also the geographic distribution of the study sample, were characterized. It was observed that the spoligopatterns family distribution was similar to that reported in South America,10,20–23 where the LAM and H lineages were the most prevalent. It was also found that 41 (35.7%) isolates that did not match any of the shared types present in SpolDB4 database. The results further showed a high rate-case among the resistant TB group as a result of transmitted and acquired resistance.
It was found a variety of family and subfamilies, which are also found in other regions of the world.18 These might reflect the effects of migratory flow and population mix in Brazil. As an example, the Haarlem lineage, which was the second-most frequent in our study, was chronologically originated in Holland. It is hypothesized that the strain was introduced in Northeast Brazil during the colonization by the Dutch in the 17th Century. In addition to this, differences were found in our study regarding some frequency lineages in comparison to other studies conducted in other regions of the country, where the T family is described as the second or third-most prevalent.9,10,21,23
Confirming to the literature,18,21,24 the LAM family was the most frequently found in this study, from which the subfamily LAM9 was the most common. However, our results differ in the frequency of the other subfamilies of LAM as compared to other studies conducted in other regions of Brazil such as the absence of LAM2. The movement of Europeans, at historical and contemporary time, might explain the presence of LAM3 lineage in Brazil.
Although not significant, the fact that some of our isolates are not commonly found in Brazil such as East African-Indian strains that is predominant in India, might be explained that some of the lineages have not adapted to spreading within a population and show a significantly low case frequency. This hypothesis is supported by some of the reports18,25,26 suggesting that certain genotypes of M. tuberculosis might be adapted to host ethnicity and geography. It is suggested that the ancestral EAI lineage shows certain characteristics, including resistance to INH.27 However, both of our isolates were INH sensitive. Despite MDR-TB outbreaks having been associated with Beijing and H families,28,29 in our study there was no isolation of Beijing strain and also no association of H family and R-TB.
It is well-reported that mostly drug-resistant TB can be generated by either acquired or amplified resistance, as a consequence of the treatment failure and abandon.30,31 In our results, despite history of previous treatment being considered as a risk factor for development of resistant TB bacilli, a total of 20 (31.2%) patients in the S-TB group reported history of previous treatment.30,32 Moreover, a high proportion of MDR-TB cases had a history of previous treatment (84.3%), due to either irregular treatment or failure, which was much higher than reported in the literature.33
Many of the reports studying drug resistance rely upon laboratory data based on specimen collection; therefore they do not identify the true MDR burden at community level. According to WHO Forth global Report, there is an estimation of 3% of MDR-TB among new cases and 20–40% among previously treated cases.2 In contrast to that report, we have found a much higher frequency of MDR-TB rate among the new cases (15.7%). A previous study conducted in Southeast Brazil has found a MDR-TB rate of 5.2% among new cases and 16.7% among previously treated cases.33 Since our samples were collected by active case-finding in a reference hospital for treatment of TB, the high MDR rate among new cases reflects the true importance of surveying this group of patients. In Fortaleza, the high rate of MDR-TB in new cases is a concern that was not identified in surveillance reports. Despite some of the reports associating certain clonal strains and transmitted TB,29,34 no association was seen among the new cases MDR-TB and the identified lineages.
The combination of spatial and genotyping data suggested ongoing neighborhood transmission within the population in this municipality. The utility of geographic information systems and spatial analysis has been reported in the literature for surveillance of TB35–38 and leprosy.8 In our study, at the district level (ensemble neighborhoods), pulmonary TB cases were found to be spread over all districts, with some concentration in the south and west of Fortaleza. Population with high-income live in the North and Northeast of Fortaleza. These concentrations may reflect the underlying heterogeneous spatial distribution of socio-economic status.
The positive association between household contacts with more than one case and TB risk has been reported in previous studies.39,40 Agreeing to these, in our study a significant association of R-TB and previous treatment was observed. Furthermore, in nine of the hot spot zones studied, there were patients who had lost follow-up or failed treatment, which resulted in household transmission. The loss of follow-up implies those patients remain contagious, resulting in an increased risk of the disease within the domicile and in the neighborhood. Patients who did not complete their treatment may reflect low level of care and poor follow-up system, which highlight the deficiency in health care programs of a low-income country.
Analyzing the geographic location of the studied TB isolates, the genotyping of the isolates and the variables related to TB (the number of household of more than one case and clinical history of the case), we suggest that the transmission is a result of clustered cases and also due to the diffusion from the nearest neighborhood. In the hot spot zones, patients were linked into groups of two or four individuals because they lived in the same household, or street, or in a nearby street. Other studies have demonstrated that TB transmission can occur by casual contact and location of the TB exposure is an important factor to be considered.41,42 This finding shows the importance of geo-referencing studies in order to identify the MDR-TB spread due to active transmission.
In conclusion, this study highlights the relevant rate of transmitted resistance among MDR-TB and a typical geographical pattern of M. tuberculosis strains in Northeast Brazil. In addition, the study also showed tuberculosis outbreaks by active transmission of bacilli among clustered cases that gives this immunocompetent population a high prevalence of TB.
A combination of clinical, spoligotyping and geospatial analysis to study resistant tuberculosis was successfully applied, thus demonstrating the utility of this strategy in tuberculosis control programs. The strain typing patterns of resistant isolates were useful in providing information about distribution and epidemiological characteristics, which can be applied to allow better follow-up of patients, and in defining strategies for prevention of the disease in prevalent areas.
Financial supportThis work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq – 410508/2006-3).
Conflict of interestAll authors declare to have no conflict of interest.
The authors are grateful for the participation of the patients as well as the support of the staff at Hospital de Messejana, Ceará. In addition, they thank Creuza L. Campelo for her help at the Central Laboratory of Public Health and Walter Correia for his help during the research.