To establish a resistance (R) surveillance program monitoring antimicrobial susceptibility patterns in Latin America (LATAM; Argentina [ARG], Brazil [BRA], Chile, Colombia [CBA], Costa Rica, Ecuador [ECU], Guatemala [GUA], Mexico [MEX], Panama [PAN], Peru, and Venezuela [VEN]).
MethodsIn 2011, 4979 organisms were collected from 11 nations (20 laboratories) for susceptibility testing in a central laboratory design. Antimicrobials were tested by CLSI methods and results interpreted by CLSI and EUCAST breakpoints. Most common Gram-positive (Staphylococcus aureus [SA, 921], other staphylococci [CoNS; 299], enterococci [218], Streptococcus pneumoniae [SPN; 182], β-haemolytic streptococci [115]) and Gram-negative (E. coli [EC; 644], Klebsiella spp. [KSP; 517], Enterobacters [272], Pseudomonas aeruginosa [PSA; 586], Acinetobacters [ACB; 494]) pathogens were analyzed against linezolid (LZD), vancomycin (VAN), tigecycline (TIG), colistin (COL), cefoperazone/sulbactam (C/S), and amikacin (AMK).
ResultsMRSA rates varied from 29% (CBA, BRA) to 79% (Peru); but LZD (MIC90, 2mg/L), TIG (MIC90, 0.12mg/L) and VAN (MIC90, 1mg/L) covered all strains. Enterococci showed a 14% VRE rate, highest in BRA and MEX; all inhibited by TIG and daptomycin, but not LZD (three non-susceptible with G2576T mutations or cfr). Penicillin-R among SPN and viridans streptococci was 51.6 and 41.1%, respectively. LZD overall R against Gram-positives was 0.3%. High ESBL rates were observed in EC (54–71%) and KSP (≥50%) from GUA, MEX and Peru, and six nations, respectively. Carbapenem-R in KSP was 9%, highest rates associated with KPC in BRA, CBA, ECU, PAN and VEN; also a NDM-1 in KSP from CBA. AMK, TIG, C/S and carbapenems were the broadest-spectrum agents tested against Enterobacteriaceae. Only COL inhibited >90% of PSA; COL and TIG (≤2mg/L) covered ≥85% of ACB.
ConclusionsLATAM nations demonstrated variable levels of antimicrobial R especially among Enterobacteriaceae (β-lactamase-mediated), PSA and ACB. MRSA (48%), VRE (14%) and multidrug-R SPN were also regional therapeutic challenges.
Recent escalations of β-lactamase-mediated resistances (extended-spectrum β-lactams [ESBL], serine carbapenamases [KPCs], OXA-series Class D enzymes, and metallo-β-lactamases [MBL]) worldwide has complicated antimicrobial therapy of important/common Gram-negative bacillary infections.1–4 Already existing resistance challenges among Gram-positive cocci (methicillin-resistant staphylococci, vancomycin-resistant enterococci [VRE] and multidrug-resistant [MDR] pneumococci) further emphasize the needs for global, regional, national and local surveillance of antimicrobial susceptibility patterns to guide empiric therapy and direct or monitor interventions.5–7 These resistant strains increase patient morbidity and mortality, as well as the cost of medical care delivery.4,7
Current surveillance programs, particularly at the global level,1–3 have concentrated on larger economically developed nations where fiscal markets and supporting regulatory agencies (USA-FDA, EMA) would recognize the value, and have the resources to sustain monitoring. In contrast, surveillance data from countries outside the major markets having faced more limited support for drug resistance monitoring, drug patent protection, prescription drug law and antimicrobial stewardship programs are more limited.4 Beginning in 2011, the Latin American (LATAM) surveillance programs (SENTRY Antimicrobial Surveillance Program and several others) administered by JMI Laboratories (North Liberty, Iowa, USA) were expanded to include sites within some countries having limited sampling support or not having significant reported statistics. This regional resistance surveillance program provides reference susceptibility test information in several areas of the world including 11 countries in LATAM including seven that are uncommonly sampled (Colombia, Costa Rica, Ecuador, Guatemala, Panama, Peru and Venezuela). Data from testing nearly 5000 clinical isolates in 2011 are presented here.
Materials and methodsNations and organisms sampledEleven countries in LATAM (20 laboratory sampling sites having 93–503 organism samples/site) were sampled with a target of ≥250 isolates per nation. These institutions were generally tertiary-care hospitals. The compliance to protocol ranged from 190 [Venezuela, 95%] to >100% for the “developed” countries. The collected organisms were isolated consecutively from various types of clinical infections (prevalence design) including bloodstream (18.8%), respiratory tract (20.1%), skin and skin structure (13.1%) as well as other or unspecified body sites. The countries (sites; sample size) were: Argentina (two; 498), Brazil (five; 1588), Chile (two; 467), Colombia (one; 208), Costa Rica (one; 193), Ecuador (one; 192), Guatemala (one; 201), Mexico (three, 1052), Panama (one; 196), Peru (one; 194) and Venezuela (two, 190); one isolate per patient per infectious episode, see Table 1. The organisms forwarded to the monitoring central laboratory (JMI Laboratories) were as follows: S. aureus (921), coagulase-negative Staphylococcus species (CoNS; 299), enterococci (218; 92.2% E. faecalis or E. faecium), S. pneumoniae (182), β-haemolytic streptococci (115; 92.2% S. pyogenes or S. agalactiae), viridans group streptococci (90; more than eight species), E. coli (644; 37.3% ESBL phenotype), Klebsiella spp. (517; three species, 52.4% ESBL phenotype), Enterobacter spp. (272), P. mirabilis (74; 24.3% ESBL phenotype), other Enterobacteriaceae (292), H. influenzae (128; 29.7% β-lactamase-positive), M. catarrhalis (33), P. aeruginosa (586), and Acinetobacter spp. (494; 94.7% A. baumannii). A total of 4979 isolates were tested, 4865 or 97.7% of which are presented in Tables 2 and 3; the remaining organisms occurred in small numbers precluding a significant sample size per species, e.g. limited analytical power.
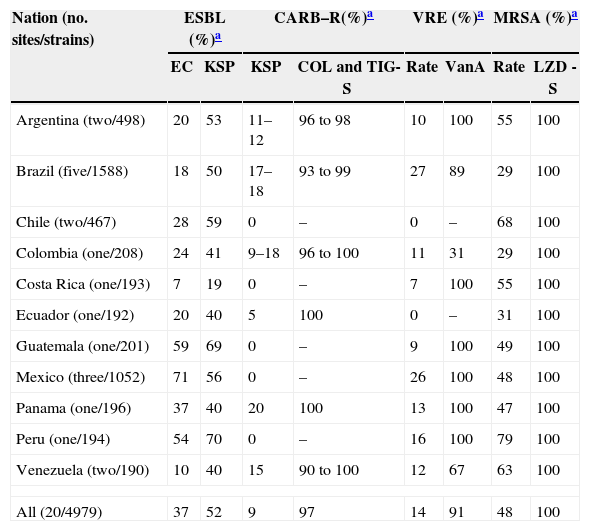
Summary of important emerging resistance profiles detected in 11 Latin American countries (20 medical centers; 2011); a 4979 isolate sample.
| Nation (no. sites/strains) | ESBL (%)a | CARB–R(%)a | VRE (%)a | MRSA (%)a | ||||
|---|---|---|---|---|---|---|---|---|
| EC | KSP | KSP | COL and TIG-S | Rate | VanA | Rate | LZD -S | |
| Argentina (two/498) | 20 | 53 | 11–12 | 96 to 98 | 10 | 100 | 55 | 100 |
| Brazil (five/1588) | 18 | 50 | 17–18 | 93 to 99 | 27 | 89 | 29 | 100 |
| Chile (two/467) | 28 | 59 | 0 | – | 0 | – | 68 | 100 |
| Colombia (one/208) | 24 | 41 | 9–18 | 96 to 100 | 11 | 31 | 29 | 100 |
| Costa Rica (one/193) | 7 | 19 | 0 | – | 7 | 100 | 55 | 100 |
| Ecuador (one/192) | 20 | 40 | 5 | 100 | 0 | – | 31 | 100 |
| Guatemala (one/201) | 59 | 69 | 0 | – | 9 | 100 | 49 | 100 |
| Mexico (three/1052) | 71 | 56 | 0 | – | 26 | 100 | 48 | 100 |
| Panama (one/196) | 37 | 40 | 20 | 100 | 13 | 100 | 47 | 100 |
| Peru (one/194) | 54 | 70 | 0 | – | 16 | 100 | 79 | 100 |
| Venezuela (two/190) | 10 | 40 | 15 | 90 to 100 | 12 | 67 | 63 | 100 |
| All (20/4979) | 37 | 52 | 9 | 97 | 14 | 91 | 48 | 100 |
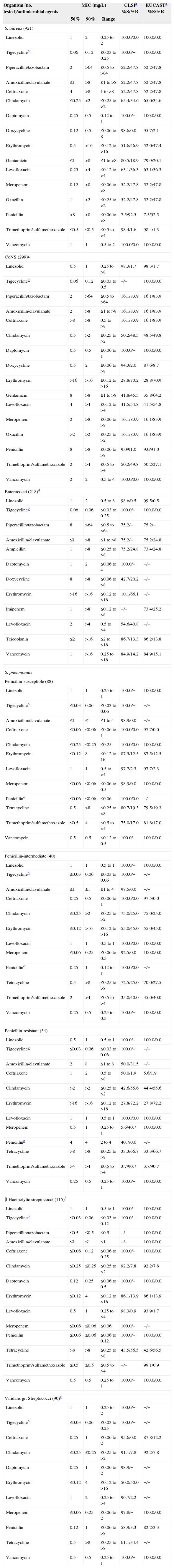
Activity of selected antimicrobial agents when tested against 1825 Gram-positive pathogens from Latin America nations (2011).
| Organism (no. tested)/antimicrobial agents | MIC (mg/L) | CLSIa %S/%R | EUCASTa %S/%R | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| S. aureus (921) | |||||
| Linezolid | 1 | 2 | 0.25 to 2 | 100.0/0.0 | 100.0/0.0 |
| Tigecyclineb | 0.06 | 0.12 | ≤0.03 to 0.25 | 100.0/− | 100.0/0.0 |
| Piperacillin/tazobactam | 2 | >64 | ≤0.5 to >64 | 52.2/47.8 | 52.2/47.8 |
| Amoxicillin/clavulanate | ≤1 | >8 | ≤1 to >8 | 52.2/47.8 | 52.2/47.8 |
| Ceftriaxone | 4 | >8 | 1 to >8 | 52.2/47.8 | 52.2/47.8 |
| Clindamycin | ≤0.25 | >2 | ≤0.25 to >2 | 65.4/34.6 | 65.0/34.6 |
| Daptomycin | 0.25 | 0.5 | 0.12 to 1 | 100.0/− | 100.0/0.0 |
| Doxycycline | 0.12 | 0.5 | ≤0.06 to 8 | 98.6/0.0 | 95.7/2.1 |
| Erythromycin | 0.5 | >16 | ≤0.12 to >16 | 51.6/46.9 | 52.0/47.4 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 80.5/18.9 | 79.9/20.1 |
| Levofloxacin | 0.25 | >4 | ≤0.12 to >4 | 63.1/36.3 | 63.1/36.3 |
| Meropenem | 0.12 | >8 | ≤0.06 to >8 | 52.2/47.8 | 52.2/47.8 |
| Oxacillin | 1 | >2 | ≤0.25 to >2 | 52.2/47.8 | 52.2/47.8 |
| Penicillin | >8 | >8 | ≤0.06 to >8 | 7.5/92.5 | 7.5/92.5 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | ≤0.5 | ≤0.5 to >4 | 98.4/1.6 | 98.4/1.3 |
| Vancomycin | 1 | 1 | 0.5 to 2 | 100.0/0.0 | 100.0/0.0 |
| CoNS (299)c | |||||
| Linezolid | 0.5 | 1 | 0.25 to >8 | 98.3/1.7 | 98.3/1.7 |
| Tigecyclineb | 0.06 | 0.12 | ≤0.03 to 0.5 | −/− | 100.0/0.0 |
| Piperacillin/tazobactam | 2 | >64 | ≤0.5 to >64 | 16.1/83.9 | 16.1/83.9 |
| Amoxicillin/clavulanate | 2 | >8 | ≤1 to >8 | 16.1/83.9 | 16.1/83.9 |
| Ceftriaxone | >8 | >8 | 0.5 to >8 | 16.1/83.9 | 16.1/83.9 |
| Clindamycin | 0.5 | >2 | ≤0.25 to >2 | 50.2/48.5 | 48.5/49.8 |
| Daptomycin | 0.5 | 0.5 | ≤0.06 to 1 | 100.0/− | 100.0/0.0 |
| Doxycycline | 0.5 | 2 | ≤0.06 to >8 | 94.3/2.0 | 87.6/8.7 |
| Erythromycin | >16 | >16 | ≤0.12 to >16 | 28.8/70.2 | 28.8/70.9 |
| Gentamicin | 8 | >8 | ≤1 to >8 | 41.8/45.5 | 35.8/64.2 |
| Levofloxacin | 4 | >4 | ≤0.12 to >4 | 41.5/54.8 | 41.5/54.8 |
| Meropenem | 2 | >8 | ≤0.06 to >8 | 16.1/83.9 | 16.1/83.9 |
| Oxacillin | >2 | >2 | ≤0.25 to >2 | 16.1/83.9 | 16.1/83.9 |
| Penicillin | 8 | >8 | ≤0.06 to >8 | 9.0/91.0 | 9.0/91.0 |
| Trimethoprim/sulfamethoxazole | 2 | >4 | ≤0.5 to >4 | 50.2/49.8 | 50.2/27.1 |
| Vancomycin | 2 | 2 | 0.5 to 4 | 100.0/0.0 | 100.0/0.0 |
| Enterococci (218)d | |||||
| Linezolid | 1 | 2 | 0.5 to 8 | 98.6/0.5 | 99.5/0.5 |
| Tigecyclineb | 0.06 | 0.06 | ≤0.03 to 0.25 | 100.0/− | 100.0/0.0 |
| Piperacillin/tazobactam | 8 | >64 | ≤0.5 to >64 | 75.2/− | 75.2/− |
| Amoxicillin/clavulanate | ≤1 | >8 | ≤1 to >8 | 75.2/− | 75.2/24.8 |
| Ampicillin | 1 | >8 | ≤0.25 to >8 | 75.2/24.8 | 73.4/24.8 |
| Daptomycin | 1 | 2 | ≤0.06 to 4 | 100.0/− | −/− |
| Doxycycline | 8 | >8 | ≤0.06 to >8 | 42.7/20.2 | −/− |
| Erythromycin | >16 | >16 | ≤0.12 to >16 | 10.1/66.1 | −/− |
| Imipenem | 1 | >8 | ≤0.12 to >8 | −/− | 73.4/25.2 |
| Levofloxacin | 2 | >4 | 0.5 to >4 | 54.6/40.8 | −/− |
| Teicoplanin | ≤2 | >16 | ≤2 to >16 | 86.7/13.3 | 86.2/13.8 |
| Vancomycin | 1 | >16 | 0.25 to >16 | 84.9/14.2 | 84.9/15.1 |
| S. pneumoniae | |||||
| Penicillin-susceptible (88) | |||||
| Linezolid | 1 | 1 | 0.25 to 1 | 100.0/− | 100.0/0.0 |
| Tigecyclineb | ≤0.03 | 0.06 | ≤0.03 to 0.06 | 100.0/− | −/− |
| Amoxicillin/clavulanate | ≤1 | ≤1 | ≤1 to 4 | 98.9/0.0 | −/− |
| Ceftriaxone | ≤0.06 | ≤0.06 | ≤0.06 to 1 | 100.0/0.0 | 97.7/0.0 |
| Clindamycin | ≤0.25 | ≤0.25 | ≤0.25 | 100.0/0.0 | 100.0/0.0 |
| Erythromycin | ≤0.12 | 8 | ≤0.12 to 16 | 87.5/12.5 | 87.5/12.5 |
| Levofloxacin | 1 | 1 | 0.5 to >4 | 97.7/2.3 | 97.7/2.3 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 to 0.5 | 98.9/0.0 | 100.0/0.0 |
| Penicilline | ≤0.06 | ≤0.06 | ≤0.06 | 100.0/0.0 | −/− |
| Tetracycline | 0.5 | >8 | ≤0.25 to >8 | 80.7/19.3 | 79.5/19.3 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | 4 | ≤0.5 to >4 | 75.0/17.0 | 81.8/17.0 |
| Vancomycin | 0.5 | 0.5 | ≤0.12 to 0.5 | 100.0/− | 100.0/0.0 |
| Penicillin-intermediate (40) | |||||
| Linezolid | 1 | 1 | 0.5 to 1 | 100.0/− | 100.0/0.0 |
| Tigecyclineb | ≤0.03 | 0.06 | ≤0.03 to 0.06 | 100.0/− | −/− |
| Amoxicillin/clavulanate | ≤1 | ≤1 | ≤1 to 4 | 97.5/0.0 | −/− |
| Ceftriaxone | 0.25 | 0.5 | ≤0.06 to 1 | 100.0/0.0 | 97.5/0.0 |
| Clindamycin | ≤0.25 | >2 | ≤0.25 to >2 | 75.0/25.0 | 75.0/25.0 |
| Erythromycin | ≤0.12 | >16 | ≤0.12 to >16 | 55.0/45.0 | 55.0/45.0 |
| Levofloxacin | 1 | 1 | 0.5 to 1 | 100.0/0.0 | 100.0/0.0 |
| Meropenem | ≤0.06 | 0.25 | ≤0.06 to 0.5 | 92.5/0.0 | 100.0/0.0 |
| Penicilline | 0.25 | 1 | 0.12 to 1 | 100.0/0.0 | −/− |
| Tetracycline | 0.5 | >8 | ≤0.25 to >8 | 72.5/25.0 | 70.0/27.5 |
| Trimethoprim/sulfamethoxazole | 2 | >4 | ≤0.5 to >4 | 35.0/40.0 | 35.0/40.0 |
| Vancomycin | 0.25 | 0.5 | 0.25 to 0.5 | 100.0/− | 100.0/0.0 |
| Penicillin-resistant (54) | |||||
| Linezolid | 0.5 | 1 | 0.5 to 1 | 100.0/− | 100.0/0.0 |
| Tigecyclineb | ≤0.03 | 0.06 | ≤0.03 to 0.06 | 100.0/− | −/− |
| Amoxicillin/clavulanate | 2 | 8 | ≤1 to 8 | 50.0/31.5 | −/− |
| Ceftriaxone | 1 | 2 | 0.5 to >8 | 50.0/1.9 | 5.6/1.9 |
| Clindamycin | >2 | >2 | ≤0.25 to >2 | 42.6/55.6 | 44.4/55.6 |
| Erythromycin | >16 | >16 | ≤0.12 to >16 | 27.8/72.2 | 27.8/72.2 |
| Levofloxacin | 1 | 1 | 0.5 to 1 | 100.0/0.0 | 100.0/0.0 |
| Meropenem | 0.5 | 1 | 0.25 to 1 | 5.6/40.7 | 100.0/0.0 |
| Penicilline | 4 | 4 | 2 to 4 | 40.7/0.0 | −/− |
| Tetracycline | >8 | >8 | ≤0.25 to >8 | 33.3/66.7 | 33.3/66.7 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 3.7/90.7 | 3.7/90.7 |
| Vancomycin | 0.25 | 0.5 | 0.25 to 1 | 100.0/− | 100.0/0.0 |
| β-Haemolytic streptococci (115)f | |||||
| Linezolid | 1 | 1 | 0.5 to 1 | 100.0/− | 100.0/0.0 |
| Tigecyclineb | ≤0.03 | 0.06 | ≤0.03 to 0.12 | 100.0/− | 100.0/0.0 |
| Piperacillin/tazobactam | ≤0.5 | ≤0.5 | ≤0.5 | −/− | 100.0/0.0 |
| Amoxicillin/clavulanate | ≤1 | ≤1 | ≤1 | −/− | 100.0/0.0 |
| Ceftriaxone | ≤0.06 | 0.12 | ≤0.06 to 0.25 | 100.0/− | 100.0/0.0 |
| Clindamycin | ≤0.25 | ≤0.25 | ≤0.25 to >2 | 92.2/7.8 | 92.2/7.8 |
| Daptomycin | 0.12 | 0.25 | ≤0.06 to 0.5 | 100.0/− | 100.0/0.0 |
| Erythromycin | ≤0.12 | 4 | ≤0.12 to >16 | 86.1/13.9 | 86.1/13.9 |
| Levofloxacin | 0.5 | 1 | 0.25 to >4 | 98.3/0.9 | 93.9/1.7 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 | 100.0/− | −/− |
| Penicillin | ≤0.06 | ≤0.06 | ≤0.06 to 0.12 | 100.0/− | 100.0/0.0 |
| Tetracycline | >8 | >8 | ≤0.25 to >8 | 43.5/56.5 | 42.6/56.5 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | ≤0.5 | ≤0.5 to >4 | −/− | 99.1/0.9 |
| Vancomycin | 0.5 | 0.5 | 0.25 to 1 | 100.0/− | 100.0/0.0 |
| Viridans gr. Streptococci (90)g | |||||
| Linezolid | 1 | 1 | 0.25 to 2 | 100.0/− | −/− |
| Tigecyclineb | ≤0.03 | 0.06 | ≤0.03 to 0.25 | 100.0/− | −/− |
| Ceftriaxone | 0.25 | 1 | ≤0.06 to 2 | 95.6/0.0 | 87.8/12.2 |
| Clindamycin | ≤0.25 | ≤0.25 | ≤0.25 to >2 | 91.1/7.8 | 92.2/7.8 |
| Daptomycin | 0.25 | 1 | ≤0.06 to 2 | 98.9/− | −/− |
| Erythromycin | ≤0.12 | 4 | ≤0.12 to >16 | 50.0/50.0 | −/− |
| Levofloxacin | 1 | 2 | 0.25 to >4 | 96.7/2.2 | −/− |
| Meropenem | ≤0.06 | 0.25 | ≤0.06 to 2 | 97.8/− | 100.0/0.0 |
| Penicillin | 0.12 | 1 | ≤0.06 to >8 | 58.9/3.3 | 82.2/3.3 |
| Tetracycline | 0.5 | >8 | ≤0.25 to >8 | 61.1/34.4 | −/− |
| Vancomycin | 0.5 | 0.5 | 0.25 to 1 | 100.0/− | 100.0/0.0 |
Criteria as published by the CLSI and EUCAST9,10, β-lactam susceptibility should be directed by the oxacillin test results.
Includes: Staphylococcus auricularis (one strain), S. capitis (10 strains), S. epidermidis (118 strains), S. equorum (one strain), S. haemolyticus (48 strains), S. hominis (29 strains), S. lugdunensis (10 strains), S.saprophyticus (six strains), S. warneri (three strains), S. xylosus (three strains), and unspeciated coagulase-negative staphylococci (70 strains).
Includes: Enterococcus avium (11 strains), E. durans (one strain), E. faecalis (142 strains), E. faecium (59 strains), E. gallinarum (four strains), and E. hirae (one strain).
Criteria were those published by the CLSI9 for ‘Penicillin parenteral (non-meningitis)’, as were the ceftriaxone breakpoints.
Includes: Streptococcus dysgalactiae (three strains), Group A Streptococcus (44 strains), Group B Streptococcus (62 strains), Group C Streptococcus (one strain), Group F Streptococcus (one strain), and Group G Streptococcus (four strains).
Includes: Streptococcus anginosus (five strains), S. bovis (one strain), S. gallolyticus (seven strains), S. infantarius (one strain), S. mitis (16 strains), S. parasanguinis (one strain), S. salivarius (two strains), S. sanguinis (two strains), unspeciated Streptococcus (one strain), and unspeciated viridans group streptococci (54 strains).
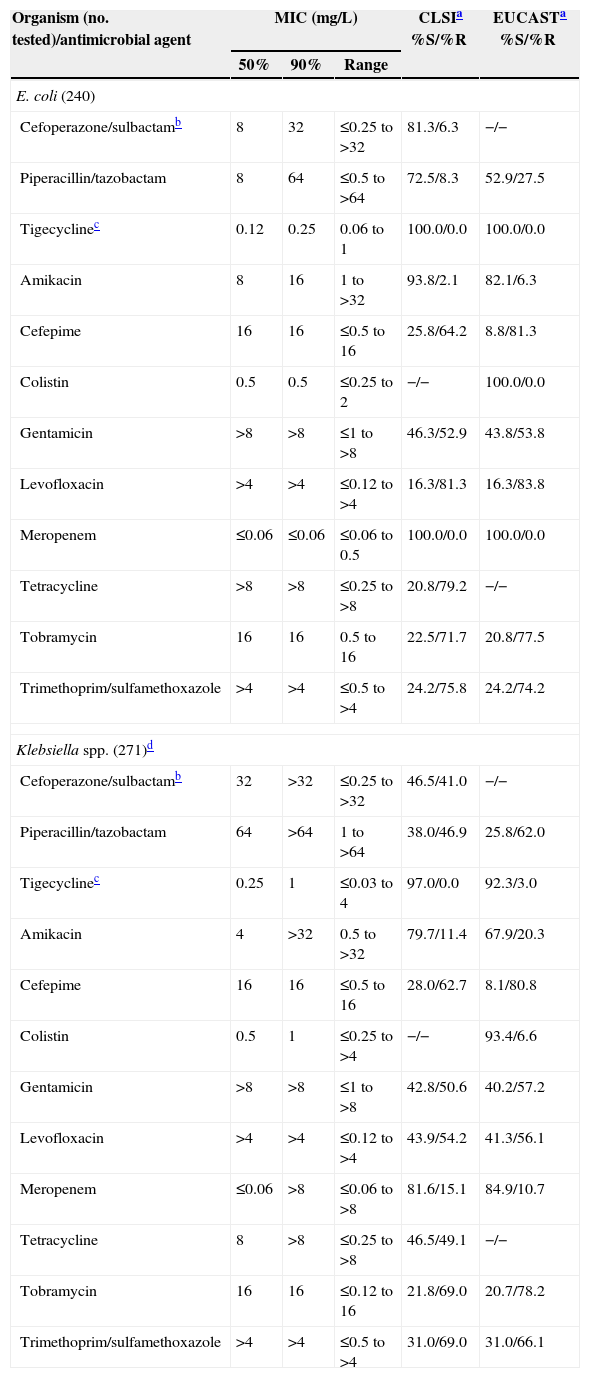
Activity of selected antimicrobial agents when tested against 3040 isolates of Gram-negative pathogens from Latin American nations (2011).
| Organism (no. tested)/antimicrobial agent | MIC (mg/L) | CLSIa %S/%R | EUCASTa %S/%R | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| E. coli (644) | |||||
| Ampicillin/sulbactam | 16 | >32 | ≤0.25 to >32 | 30.4/49.1 | 30.4/69.6 |
| Cefoperazone | 4 | >32 | ≤0.25 to >32 | 59.6/38.2 | −/− |
| Cefoperazone/sulbactamb | 2 | 16 | ≤0.25 to >32 | 92.7/2.3 | −/− |
| Piperacillin/tazobactam | 2 | 32 | ≤0.5 to >64 | 86.5/5.0 | 78.7/13.5 |
| Tigecyclinec | 0.12 | 0.25 | ≤0.03 to 1 | 100.0/0.0 | 100.0/0.0 |
| Amikacin | 4 | 8 | 0.5 to >32 | 97.5/0.8 | 92.4/2.5 |
| Amoxicillin/clavulanate | 8 | >8 | ≤1 to >8 | 58.4/41.6 | 58.4/41.6 |
| Ampicillin | >8 | >8 | 1 to >8 | 23.3/76.7 | 23.3/76.7 |
| Cefepime | ≤0.5 | >16 | ≤0.5 to >16 | 72.4/23.9 | 64.8/30.4 |
| Ceftazisdime | 0.25 | 32 | 0.03 to >32 | 69.6/27.2 | 65.5/30.4 |
| Ceftriaxone | 0.12 | >8 | ≤0.06 to >8 | 62.9/37.1 | 62.9/37.1 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 72.4/27.0 | 70.7/27.6 |
| Levofloxacin | 4 | >4 | ≤0.12 to >4 | 49.7/47.2 | 49.4/50.3 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 to 0.5 | 100.0/0.0 | 100.0/0.0 |
| Tetracycline | >8 | >8 | ≤0.25 to >8 | 40.4/59.3 | −/− |
| Tobramycin | 1 | >16 | 0.25 to >16 | 62.6/32.3 | 61.2/37.4 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 40.0/60.0 | 40.0/59.3 |
| Klebsiella spp. (517)d | |||||
| Ampicillin/sulbactam | 32 | >32 | 1 to >32 | 40.6/53.6 | 40.6/59.4 |
| Cefoperazone | >32 | >32 | ≤0.25 to >32 | 47.2/51.1 | −/− |
| Cefoperazone/sulbactamd | 4 | >32 | ≤0.25 to >32 | 71.8/21.5 | −/− |
| Piperacillin/tazobactam | 4 | >64 | ≤0.5 to >64 | 66.2/25.1 | 59.0/33.8 |
| Tigecyclinec | 0.25 | 1 | ≤0.03 to >4 | 97.9/0.2 | 95.0/2.1 |
| Amikacin | 2 | 32 | 0.5 to >32 | 89.0/6.0 | 82.8/11.0 |
| Amoxicillin/clavulanate | 8 | >8 | ≤1 to >8 | 52.4/47.6 | 52.4/47.6 |
| Cefepime | 1 | >16 | ≤0.5 to >16 | 62.3/32.9 | 51.8/42.4 |
| Ceftazidime | 1 | >32 | ≤0.015 to >32 | 57.3/37.3 | 51.8/42.7 |
| Ceftriaxone | 8 | >8 | ≤0.06 to >8 | 48.7/51.1 | 48.7/51.1 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 68.5/27.5 | 67.1/31.5 |
| Levofloxacin | 0.25 | >4 | ≤0.12 to >4 | 68.7/30.0 | 67.1/31.3 |
| Meropenem | ≤0.06 | 1 | ≤0.06 to >8 | 90.3/7.9 | 92.1/5.6 |
| Tetracycline | 2 | >8 | ≤0.25 to >8 | 62.3/35.2 | −/− |
| Tobramycin | 1 | >16 | ≤0.12 to 16 | 57.6/37.1 | 56.9/42.4 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | >4 | ≤0.5 to >4 | 57.1/42.9 | 57.1/41.0 |
| P. mirabilis (74) | |||||
| Ampicillin/sulbactam | 2 | 32 | 0.5 to 32 | 78.4/10.8 | 78.4/21.6 |
| Cefoperazone | 1 | >32 | ≤0.25 to >32 | 74.3/20.3 | −/− |
| Cefoperazone/sulbactamb | 1 | 4 | ≤0.25 to 16 | 100.0/0.0 | −/− |
| Piperacillin/tazobactam | ≤0.5 | 1 | ≤0.5 to 2 | 100.0/0.0 | 100.0/0.0 |
| Tigecyclinec | 2 | 4 | 0.5 to >4 | 85.1/1.4 | 32.4/14.9 |
| Amikacin | 4 | 8 | 1 to >32 | 95.9/2.7 | 90.5/4.1 |
| Amoxicillin/clavulanate | ≤1 | 8 | ≤1 to >8 | 93.2/6.8 | 93.2/6.8 |
| Ampicillin | 2 | >8 | 0.5 to >8 | 52.7/47.3 | 52.7/47.3 |
| Cefepime | ≤0.5 | >16 | ≤0.5 to 16 | 81.1/17.6 | 75.7/20.3 |
| Ceftazidime | 0.06 | 2 | 0.03 to >32 | 94.6/5.4 | 87.8/5.4 |
| Ceftriaxone | ≤0.06 | >8 | ≤0.06 to >8 | 75.7/23.0 | 75.7/23.0 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 78.4/21.6 | 75.7/21.6 |
| Imipenem | 1 | 2 | ≤0.12 to 4 | 73.0/4.1 | 95.9/0.0 |
| Levofloxacin | ≤0.12 | >4 | ≤0.12 to >4 | 73.0/23.0 | 67.6/27.0 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 to 0.12 | 100.0/0.0 | 100.0/0.0 |
| Tobramycin | 1 | 16 | 0.5 to 16 | 77.0/12.2 | 73.0/23.0 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 47.3/52.7 | 47.3/51.4 |
| Enterobacter spp. (272)e | |||||
| Cefoperazone | 2 | >32 | ≤0.25 to >32 | 59.9/34.9 | −/− |
| Cefoperazone/sulbactamb | 1 | 32 | ≤0.25 to >32 | 84.9/6.3 | −/− |
| Piperacillin/tazobactam | 4 | >64 | ≤0.5 to >64 | 75.7/10.7 | 69.5/24.3 |
| Tigecyclinec | 0.25 | 1 | 0.06 to 4 | 97.8/0.0 | 94.1/2.2 |
| Amikacin | 2 | 16 | 0.5 to >32 | 94.1/4.0 | 86.8/5.9 |
| Cefepime | ≤0.5 | >16 | ≤0.5 to >16 | 84.6/12.1 | 70.2/21.0 |
| Ceftazidime | 0.5 | >32 | 0.06 to >32 | 63.2/33.5 | 57.7/36.8 |
| Ceftriaxone | 0.5 | >8 | ≤0.06 to >8 | 55.5/44.5 | 55.5/44.5 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 77.9/19.5 | 76.5/22.1 |
| Levofloxacin | ≤0.12 | >4 | ≤0.12 to >4 | 80.9/16.5 | 79.4/19.1 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06 to >8 | 98.2/1.5 | 98.5/0.4 |
| Tetracycline | 2 | >8 | 0.5 to >8 | 74.3/19.1 | −/− |
| Tobramycin | 0.5 | >16 | ≤0.12 to >16 | 69.9/28.3 | 69.5/30.1 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | >4 | ≤0.5 to >4 | 71.0/29.0 | 71.0/28.3 |
| Indole-positive Proteus spp. (94)f | |||||
| Cefoperazone | 4 | >32 | ≤0.25 to >32 | 71.3/21.3 | −/− |
| Cefoperazone/sulbactamb | 2 | 8 | ≤0.25 to 32 | 98.9/0.0 | −/− |
| Piperacillin/tazobactam | ≤0.5 | 4 | ≤0.5 to >64 | 98.9/1.1 | 97.9/1.1 |
| Tigecyclinec | 0.5 | 2 | 0.25 to 4 | 94.7/0.0 | 89.4/5.3 |
| Amikacin | 2 | 8 | 0.5 to >32 | 94.7/4.3 | 92.6/5.3 |
| Cefepime | ≤0.5 | 16 | ≤0.5 to >16 | 89.4/4.3 | 80.9/13.8 |
| Ceftazidime | 0.25 | 16 | 0.03 to >32 | 80.9/13.8 | 70.2/19.1 |
| Ceftriaxone | 0.12 | >8 | ≤0.06 to >8 | 68.1/25.5 | 68.1/25.5 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 68.1/27.7 | 61.7/31.9 |
| Imipenem | 2 | 2 | 0.25 to 4 | 35.1/6.4 | 93.6/0.0 |
| Levofloxacin | 1 | >4 | ≤0.12 to >4 | 62.8/25.5 | 55.3/37.2 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06 to 1 | 100.0/0.0 | 100.0/0.0 |
| Tobramycin | 1 | 16 | 0.25 to >16 | 73.4/14.9 | 68.1/26.6 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 43.6/56.4 | 43.6/56.4 |
| Serratia spp. (142)g | |||||
| Cefoperazone | 2 | >32 | 0.5 to >32 | 78.9/14.1 | −/− |
| Cefoperazone/sulbactamb | 2 | 16 | 0.5 to >32 | 90.8/4.9 | −/− |
| Piperacillin/tazobactam | 2 | 32 | ≤0.5 to >64 | 89.4/7.0 | 85.9/10.6 |
| Tigecyclinec | 0.5 | 1 | 0.25 to >4 | 95.8/0.7 | 90.1/4.2 |
| Amikacin | 2 | 16 | 0.5 to >32 | 90.8/5.6 | 85.9/9.2 |
| Cefepime | ≤0.5 | 4 | ≤0.5 to >16 | 92.3/7.0 | 84.5/9.2 |
| Ceftazidime | 0.12 | 16 | 0.06 to >32 | 84.5/13.4 | 81.0/15.5 |
| Ceftriaxone | 0.25 | >8 | ≤0.06 to >8 | 75.4/23.2 | 75.4/23.2 |
| Gentamicin | ≤1 | >8 | ≤1 to >8 | 85.2/13.4 | 83.1/14.8 |
| Levofloxacin | ≤0.12 | 4 | ≤0.12 to >4 | 88.7/7.7 | 83.1/11.3 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06 to 4 | 98.6/0.7 | 99.3/0.0 |
| Tobramycin | 4 | >16 | 0.25 to >16 | 73.9/19.7 | 45.8/26.1 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | >4 | ≤0.5 to >4 | 84.5/15.5 | 84.5/12.7 |
| Citrobacter spp. (56)h | |||||
| Cefoperazone | 1 | >32 | ≤0.25 to >32 | 69.6/25.0 | −/− |
| Cefoperazone/sulbactamb | 0.5 | 16 | ≤0.25 to >32 | 91.1/7.1 | −/− |
| Piperacillin/tazobactam | 4 | 64 | 1 to >64 | 80.4/5.4 | 76.8/19.6 |
| Tigecyclinec | 0.25 | 0.5 | 0.06 to 2 | 100.0/0.0 | 96.4/0.0 |
| Amikacin | 2 | 32 | 0.5 to >32 | 89.3/5.4 | 83.9/10.7 |
| Cefepime | ≤0.5 | 16 | ≤0.5 to 16 | 87.5/10.7 | 76.8/16.1 |
| Ceftazidime | 0.5 | >32 | 0.06 to >32 | 71.4/25.0 | 66.1/28.6 |
| Ceftriaxone | 0.25 | >8 | ≤0.06 to >8 | 66.1/32.1 | 66.1/32.1 |
| Gentamicin | ≤1 | 4 | ≤1 to >8 | 91.1/8.9 | 83.9/8.9 |
| Levofloxacin | ≤0.12 | 1 | ≤0.12 to >4 | 92.9/5.4 | 92.9/7.1 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 to 4 | 98.2/1.8 | 98.2/0.0 |
| Tetracycline | 1 | 4 | 1 to >8 | 91.1/8.9 | −/− |
| Tobramycin | 1 | 16 | 0.25 to 16 | 80.4/19.6 | 78.6/19.6 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | >4 | ≤0.5 to >4 | 76.8/23.2 | 76.8/23.2 |
| H. influenzae (128) | |||||
| Piperacillin/tazobactam | ≤0.5 | ≤0.5 | ≤0.5 | 100.0/0.0 | −/− |
| Tigecyclinec | 0.25 | 0.5 | 0.06 to 1 | 86.7/- | −/− |
| Amoxicillin/clavulanate | ≤1 | 2 | ≤1 to 2 | 100.0/0.0 | 100.0/0.0 |
| Ampicillin | 0.25 | >8 | ≤0.12 to >8 | 70.3/28.9 | 70.3/29.7 |
| Cefepime | ≤0.5 | ≤0.5 | ≤0.5 | 100.0/− | 100.0/0.0 |
| Ceftriaxone | ≤0.06 | ≤0.06 | ≤0.06 to 0.5 | 100.0/− | 99.2/0.8 |
| Levofloxacin | ≤0.12 | ≤0.12 | ≤0.12 | 100.0/− | 100.0/0.0 |
| Meropenem | ≤0.06 | 0.12 | ≤0.06 to 0.25 | 100.0/− | 100.0/0.0 |
| Tetracycline | 0.5 | 0.5 | ≤0.12 to 16 | 98.4/1.6 | 98.4/1.6 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | >4 | ≤0.5 to >4 | 61.7/35.2 | 61.7/37.5 |
| M. catarrhalis (33) | |||||
| Tigecyclinec | 0.06 | 0.06 | 0.03 to 0.06 | −/− | −/− |
| Amoxicillin/clavulanate | ≤1 | ≤1 | ≤1 | 100.0/0.0 | 100.0/0.0 |
| Cefepime | 1 | 2 | ≤0.5 to 4 | −/− | 100.0/0.0 |
| Ceftriaxone | 0.25 | 0.5 | ≤0.06 to 0.5 | 100.0/− | 100.0/0.0 |
| Levofloxacin | ≤0.12 | ≤0.12 | ≤0.12 to 1 | 100.0/− | 100.0/0.0 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 | −/− | 100.0/0.0 |
| Tetracycline | 0.25 | 0.25 | ≤0.12 to 0.5 | 100.0/0.0 | 100.0/0.0 |
| Trimethoprim/sulfamethoxazole | ≤0.5 | ≤0.5 | ≤0.5 | 100.0/0.0 | 100.0/0.0 |
| P. aeruginosa (586) | |||||
| Cefoperazoneb | 32 | >32 | 0.5 to >32 | 49.3/39.4 | −/− |
| Cefoperazone/sulbactam | 16 | >32 | 0.5 to >32 | 55.8/25.4 | −/− |
| Piperacillin/tazobactam | 16 | >64 | ≤0.5 to >64 | 58.5/22.9 | 58.5/41.5 |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 75.4/20.5 | 71.3/24.6 |
| Cefepime | 8 | 16 | ≤0.5 to 16 | 63.8/25.9 | 63.8/36.2 |
| Ceftazidime | 4 | >32 | 0.25 to >32 | 65.7/29.4 | 65.7/34.3 |
| Colistin | 1 | 2 | ≤0.25 to 4 | 99.5/0.0 | 99.5/0.5 |
| Gentamicin | 2 | >8 | ≤1 to >8 | 67.4/29.4 | 67.4/32.6 |
| Imipenem | 2 | >8 | ≤0.12 to >8 | 52.9/44.9 | 55.1/28.5 |
| Levofloxacin | 2 | >4 | ≤0.12 to >4 | 56.8/38.2 | 47.8/43.2 |
| Meropenem | 2 | >8 | ≤0.06 to >8 | 54.4/38.4 | 54.4/28.2 |
| Tobramycin | 0.5 | 16 | ≤0.12 to 16 | 70.1/29.0 | 70.1/29.9 |
| Acinetobacter spp. (494)i | |||||
| Cefoperazone/sulbactam | 16 | 32 | ≤0.25 to >32 | 59.3/8.1 | −/− |
| Tigecycline | 1 | 4 | ≤0.03 to >4 | −/− | −/− |
| Amikacin | >32 | >32 | 0.5 to >32 | 25.3/67.6 | 23.1/74.7 |
| Colistin | 0.5 | 2 | ≤0.25 to >4 | 98.0/2.0 | 98.0/2.0 |
| Doxycycline | 1 | >8 | ≤0.06 to >8 | 80.4/18.6 | −/− |
| Gentamicin | >8 | >8 | ≤1 to >8 | 29.2/58.9 | 29.2/70.9 |
| Imipenem | >8 | >8 | ≤0.12 to >8 | 22.9/75.7 | 22.5/75.7 |
| Meropenem | >8 | >8 | ≤0.06 to >8 | 23.1/75.5 | 21.9/75.5 |
| Tetracycline | 8 | >8 | 0.5 to >8 | 27.3/43.3 | −/− |
| Tobramycin | 16 | 16 | 0.25 to 16 | 47.8/51.6 | 47.8/52.2 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 22.1/77.9 | 22.1/75.3 |
Includes: Klebsiella oxytoca (51 strains), K. ozaenae (two strains), K. pneumoniae (460 strains), and unspeciated Klebsiella (four strains).
Includes: Enterobacter aerogenes (47 strains), E. cloacae (202 strains), E. gergoviae (two strains), and unspeciated Enterobacter (21 strains).
Includes: Morganella morganii (72 strains), Proteus vulgaris (12 strains), P. rettgeri (five strains), P. stuartii (four strains), and unspeciated Providencia (one strain).
Includes: Serratia liquefaciens (one strain), S. marcescens (131 strains), and unspeciated Serratia (10 strains).
Organisms detected with resistances to key, available agents were tested by various molecular methods such as PCR amplification/sequencing, example ESBLs, MBLs, MDR Gram-negative bacilli or Gram-positive cocci.1,2
Methods and antimicrobials testedCLSI M07-A9 (2012) methods were applied using validated broth microdilution panels produced by ThermoFisher Scientific Inc., formerly TREK Diagnostics (Cleveland, Ohio, USA).8 Interpretations of results utilized CLSI (M100-S23, 2013), USA-Food and Drug Administration (FDA) and EUCAST (2013) criteria;9–11 and the results of quality control (QC) tests were dominantly (nearly 99.0%) within QC ranges (CLSI M100-S23) for six utilized control organisms.
The sponsor's (Pfizer Inc., New York, New York, USA) compounds included: linezolid, tigecycline, piperacillin/tazobactam, ampicillin/sulbactam, cefoperazone and cefoperazone/sulbactam. For studying Gram-negative bacilli, Gram-positive cocci, and fastidious respiratory tract species, numerous additional (15–25) drugs were also tested. ESBL patterns were defined for E. coli, Klebsiella spp. and Proteus mirabilis per CLSI (2013) criteria as a MIC of ≥2mg/L for aztreonam or ceftriaxone or ceftazidime.9,10 Carbapenem-resistant Enterobacteriaceae (CRE) were detected by a MIC at ≥2mg/L for doripenem or imipenem or meropenem.9
Results and discussionAntimicrobial profiles of 1825 Gram-positive pathogens (Tables 1 and 2)S. aureus isolates (921, 47.8% MRSA overall) exhibited complete (100.0%) susceptibility to linezolid (MIC50/90, 1/2mg/L), daptomycin (MIC50/90, 0.25/0.5mg/L), tigecycline (MIC50/90, 0.06/0.12mg/L) and vancomycin (MIC50/90, 1/1mg/L). Rare (1.1%) resistance to trimethoprim/sulfamethoxazole (TMP/SMX) was observed (Table 2). Aminoglycoside (gentamicin) resistance was approximately 20.0% with higher rates documented in Peru (72.2%), Chile (30.0%), Argentina (30.7%) and Venezuela (30.6%).
CoNS samples (299; 83.9% methicillin-resistant) showed common co-resistances and only four agents with >90% susceptibility rates including linezolid, daptomycin, doxycycline, and vancomycin (94.3–100.0% susceptible). The rare occurrences of linezolid non-susceptibility (1.7%) occurred in Brazil (five strains [4.8%]; three species [S. epidermidis, three clonal isolates with a G2576 mutation; one S. hominis with a G2576, L3 at F1475 and M156T, and L4 at 577T mutations and one S. lugdunensis with a G2576 mutation]) with MIC values of 8–32mg/L; and Mexico (two strains of S. epidermidis and S. haemolyticus having cfr±L3 or L4 mutations) with MIC values at only 4mg/L. Teicoplanin non-susceptible results (11.4% by EUCAST breakpoints) were found in Brazil (10 strains, 9.6%), Costa Rica (six strains, 42.9%), Mexico (eight strains, 8.9%), Panama (two strains, 15.4%), Peru (two strains, 14.3%), and Venezuela (five strains, 45.5%).
Enterococci (218, either E. faecalis or E. faecium) had a VRE rate of 14.2–15.1% and 91.4–93.7% with a VAN-A pattern (Tables 1 and 2). Ten nations had documented VRE (range, 7.1% [Costa Rica] to 25.7–26.5% [Brazil and Mexico]), and the best tested agents (% susceptible) were linezolid (98.6%), daptomycin (100.0%), teicoplanin (86.2–86.7%) and vancomycin (84.9%). Linezolid non-susceptibility was detected in Brazil (2.9% prevalence overall; G2576 mutations in a clonal E. faecalis) and in Panama City, Panama (13.3% prevalence; cfr clonal occurrences in E. faecalis).
S. pneumoniae (182) isolates from LATAM were dominantly penicillin-non-susceptible (51.6%; using CLSI non-meningitis breakpoints) with highest rates observed in Mexico (84.8%) and Venezuela (81.2%). Similarly, ceftriaxone non-susceptible rates were elevated (21.1–43.7%) in the same two nations. Poor coverage (low susceptible %) were noted for erythromycin (62.6%), tetracycline (63.7–64.8%) and TMP/SMX (45.1–48.4%). The best antimicrobials tested against pneumococci were levofloxacin, linezolid, tigecycline and vancomycin, each inhibiting all strains at published breakpoints (Table 2). For other streptococci, important resistance profiles observed were: (1) 13.9 and 56.5% non-susceptible for macrolides and tetracyclines in β-haemolytic streptococci, respectively, (2) ≥91.1% susceptible rates for all drugs except penicillin (58.9%, CLSI criteria), erythromycin (50.0%) and tetracycline (61.1%) in viridans group streptococci, and (3) rare daptomycin (1.1%) and fluoroquinolone non-susceptible (1.7–3.3%) rates were observed (Table 2).
Antimicrobial profiles of Gram-negative bacilli are found in Tables 3 and 4E. coli (644) had an ESBL-phenotype rate of 37.3%, see Table 4. The most active tested agents were amikacin (92.7% susceptible), cefoperazone/sulbactam (92.7%), meropenem (100.0%) and tigecycline (100.0%). The most active cephalosporin against E. coli was cefepime at 72.4% by CLSI breakpoints (Table 3). Klebsiella spp. (517) showed very elevated resistance rates (Table 3), with only four drugs inhibiting ≥80.0% of isolates (tigecycline [97.9%], colistin [96.5%], meropenem [90.3%] and amikacin [89.0%]). The ESBL phenotype rate was 52.4% (Table 4), and CRE were identified (no./percentage) in Argentina (6/10.7), Brazil (31/17.3), Colombia (4/18.2), Ecuador (2/10.0), Mexico (1/1.1), Panama (4/20.0) and Venezuela (3/15.0). The following carbapenemases were identified: KPC-2 (Brazil,3 Ecuador,2 Venezuela3), KPC-3 (Colombia,2 Panama3) and NDM-1 (Colombia1). P. mirabilis (74) showed an ESBL-phenotype rate at 24.3% and several UTI-targeted antimicrobials (ampicillin and TMP/SMX) were only 47.3–52.7% effective in vitro.
Activity of 12 antimicrobial agents when tested against ESBL-phenotype Escherichia coli and Klebsiella spp. isolated in Latin American medical centers (511 strains cultured in 2011).
| Organism (no. tested)/antimicrobial agent | MIC (mg/L) | CLSIa %S/%R | EUCASTa %S/%R | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| E. coli (240) | |||||
| Cefoperazone/sulbactamb | 8 | 32 | ≤0.25 to >32 | 81.3/6.3 | −/− |
| Piperacillin/tazobactam | 8 | 64 | ≤0.5 to >64 | 72.5/8.3 | 52.9/27.5 |
| Tigecyclinec | 0.12 | 0.25 | 0.06 to 1 | 100.0/0.0 | 100.0/0.0 |
| Amikacin | 8 | 16 | 1 to >32 | 93.8/2.1 | 82.1/6.3 |
| Cefepime | 16 | 16 | ≤0.5 to 16 | 25.8/64.2 | 8.8/81.3 |
| Colistin | 0.5 | 0.5 | ≤0.25 to 2 | −/− | 100.0/0.0 |
| Gentamicin | >8 | >8 | ≤1 to >8 | 46.3/52.9 | 43.8/53.8 |
| Levofloxacin | >4 | >4 | ≤0.12 to >4 | 16.3/81.3 | 16.3/83.8 |
| Meropenem | ≤0.06 | ≤0.06 | ≤0.06 to 0.5 | 100.0/0.0 | 100.0/0.0 |
| Tetracycline | >8 | >8 | ≤0.25 to >8 | 20.8/79.2 | −/− |
| Tobramycin | 16 | 16 | 0.5 to 16 | 22.5/71.7 | 20.8/77.5 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 24.2/75.8 | 24.2/74.2 |
| Klebsiella spp. (271)d | |||||
| Cefoperazone/sulbactamb | 32 | >32 | ≤0.25 to >32 | 46.5/41.0 | −/− |
| Piperacillin/tazobactam | 64 | >64 | 1 to >64 | 38.0/46.9 | 25.8/62.0 |
| Tigecyclinec | 0.25 | 1 | ≤0.03 to 4 | 97.0/0.0 | 92.3/3.0 |
| Amikacin | 4 | >32 | 0.5 to >32 | 79.7/11.4 | 67.9/20.3 |
| Cefepime | 16 | 16 | ≤0.5 to 16 | 28.0/62.7 | 8.1/80.8 |
| Colistin | 0.5 | 1 | ≤0.25 to >4 | −/− | 93.4/6.6 |
| Gentamicin | >8 | >8 | ≤1 to >8 | 42.8/50.6 | 40.2/57.2 |
| Levofloxacin | >4 | >4 | ≤0.12 to >4 | 43.9/54.2 | 41.3/56.1 |
| Meropenem | ≤0.06 | >8 | ≤0.06 to >8 | 81.6/15.1 | 84.9/10.7 |
| Tetracycline | 8 | >8 | ≤0.25 to >8 | 46.5/49.1 | −/− |
| Tobramycin | 16 | 16 | ≤0.12 to 16 | 21.8/69.0 | 20.7/78.2 |
| Trimethoprim/sulfamethoxazole | >4 | >4 | ≤0.5 to >4 | 31.0/69.0 | 31.0/66.1 |
Among other enteric bacilli, Enterobacter spp. showed a CRE rate at 2.9% with higher rates in Colombia and Venezuela (10.0–12.5%). Amikacin, cefoperazone/sulbactam, cefepime, carbapenems and tigecycline were quite active against these species, as were nearly all tested agents versus H. influenzae (128) and M. catarrhalis (33); see Table 3.
P. aeruginosa (586) were most susceptible to amikacin (75.4%), tobramycin (70.1%) and colistin (99.5%; Table 3). Carbapenem resistance was high due to endemic β-lactamase (SPM-1, usually in Brazil), but the most elevated rates were noted in Guatemala (75.8%), Peru (62.5–68.8%) and Ecuador (55.6%). The most active β-lactam was ceftazidime (65.7%, MIC50 at 4mg/L). Acinetobacter spp. (494, four species) were significantly inhibited (% susceptible) only by colistin (98.6%), cefoperazone/sulbactam (59.3%), doxycycline (80.4%) and tigecycline (MIC90, 4mg/L). All carbapenems, aminoglycosides and ampicillin/sulbactam showed susceptibility rates at <50%, many <20%; see Table 3.
Monitoring of nearly 5000 LATAM pathogens in 2011 documents increasing antimicrobial resistances among nearly all sampled species (Tables 1–3), confirming earlier reports.1,3,6 Although methicillin resistance was elevated among staphylococci (47.8–83.9%), several agents (daptomycin, glycopeptides, linezolid and tigecycline) retained potent activity in LATAM like elsewhere in the world.4–7 VRE appears to be expanding (14.2–15.1%, in 10 nations) as are non-susceptible rates for β-lactams in S. pneumoniae. In contrast, USA rates of VRE particularly among bacteremia isolates of E. faecium have escalated to more than 80%,12 and ceftriaxone non-susceptible rates were at 12.5% in 2009 samples of pneumococci.13 Rare linezolid-resistant (<1.0% overall) CoNS and enterococci were noted with cfr and target site mutations, as previously noted in Mexico.6
β-Lactamase-mediated (ESBL, MBL [NDM-1], Class A and D carbapenamases) resistance in E. coli, Klebsiella spp., some other Enterobacteriaceae, and non-fermentative bacilli continues to evolve (Table 3) to levels of 37.3–52.4% and few drugs have ≥90.0% level inhibition at published breakpoints.1–4,14 This demands routine use of combination empiric therapies directed by surveillance programs and patient isolate tests for LATAM patients; and interventions will be required to control further resistance escalation in this geographic region.
Conflicts of interestThe authors declare no conflicts of interest.
This study was sponsored by Pfizer. RNJ and M Castanheira are employees of JMI Laboratories, who were paid consultants to Pfizer in connection with the development of this manuscript. Co-authors have no other conflicts to disclose except for M Cepparulo who is an employee of Pfizer.