To evaluate multiplex allele specific polymerase chain reaction as a rapid molecular tool for detecting multidrug-resistant tuberculosis.
MethodsBased on drug susceptibility testing, 103 isolates were multidrug-resistant tuberculosis and 45 isolates were sensitive to isonicotinylhydrazine and rifampin. Primers were designed to target five mutations hotspots that confer resistance to the first-line drugs isoniazid and rifampin, and multiplex allele specific polymerase chain reaction was performed. Whole-genome sequencing confirmed drug resistance mutations identified by multiplex allele specific polymerase chain reaction.
ResultsDNA sequencing revealed that 68.9% of multidrug-resistant strains have point mutations at codon 315 of the katG gene, 19.8% within the mabA-inhA promoter, and 98.0% at three hotspots within rpoB. Multiplex allele specific polymerase chain reaction detected each of these five mutations, yielding 82.3% sensitivity and 100% specificity for isoniazid resistance, and 97.9% sensitivity and 100% specificity for rifampin resistance as compared to drug susceptibility testing.
ConclusionsThe results show that multiplex allele specific polymerase chain reaction is an inexpensive and practical method for rapid detection of multidrug-resistant tuberculosis in developing countries.
Tuberculosis infection is a global problem due to the high-risk of human transmission, morbidity and mortality. Nearly two million deaths and more than 9 million new cases are reported each year, making Mycobacterium tuberculosis the primary cause of death from a single pathogen.1 The spreading of HIV/AIDS and the emergence of the multidrug-resistant strains of TB (MDR-TB) led to a worldwide resurgence of tuberculosis2 which poses a serious threat to the global prevention and control of this disease. The treatment of MDR-TB requires the use of second-line drugs. However, these drugs may be of minimal therapeutic value, can be more expensive and of greater toxicity.3
A single gene mutation can induce TB drug resistance.4–6 Studies about the frequency of various gene mutations leading to MDR-TB in our region have not been conducted yet. Because these specific mutations are related to changes in TB susceptibility to certain drugs, the study of the MDR-TB mutation spectrum in our local area is helpful to guide the treatment of tuberculosis and can inform the development of a rapid molecular diagnostic test to detect variants of MDR-TB.7 The prevalent point mutations conferring drug resistance to this pathogen are: mutations in the KatG gene that allow isoniazid or isonicotinylhydrazine (INH) tolerance; mutations of the mabA-inhA operon promoter, rpoB or rifampin (RIF) tolerance determining region (RRDR) regions, which induce RIF tolerance.8
As treatment of MDR-TB is difficult and its transmission is rapid, the need for developing a rapid diagnostic test to detect MDR-TB and effectively prevent the spread of MDR-TB in the community is paramount to avoid an epidemic. The gold standard for diagnosing MDR-TB is culture-based drug susceptibility testing (DST). However, the long replication time of M. tuberculosis delays the time to reach a diagnosis via DST, and performing DST requires appropriate laboratory equipment and skilled personnel, making it difficult to adapt DST as a routine lab test. Drug resistance is determined clinically by observation of non-responsiveness to standard first-line drug therapy and through information of history of exposure to MDR-TB. Multiple allele specific PCR (MAS-PCR) allows simultaneous detection of the most common INH and RIF tolerance related gene mutations, which reduces the cost and the dependence upon technical skills.9
In the present study, we identified the most common INH and RIF mutations from previously collected clinical isolates of MDR-TB using MAS-PCR. Our results show that this method is capable of detecting those mutations quickly and economically, making this method suitable for application in developing countries.
Materials and methodsIsolation of M. tuberculosisA total of 191 M. tuberculosis clinical isolates were collected from 2005 to 2012 at our hospital. DST analysis confirmed that 103 of these isolates were MDR-TB and 45 isolates were sensitive to INH and RIF. MAS-PCR was performed using these TB clinical isolates. This study was conducted in accordance with the declaration of Helsinki and approved by the Ethics Committee of the First Affiliated Hospital of Xinxiang Medical University. Written informed consent was obtained from all participants.
Identification and drug sensitivity testsSputum samples were decontaminated according to the revised Petroff method10 and cultured on Lowenstein–Jensen agar. AccuProbe hybridization test (Gen-Probe) was used to confirm that the clones represent M. tuberculosis complex. Biochemical tests, including nitrate reduction, niacin test and 68°C catalase inhibition were conducted to further classify these isolates.10 DST was performed using the Canetti multiple proportional dilution method. Minimum inhibitory drug concentrations used were 0.2mg/mL INH, 40mg/mL RIF, 4mg/mL streptomycin and 2mg/mL ethambutol.
Extraction of genomic DNAMycobacterial genomic DNA was extracted from cultured isolates using the genomic DNA extraction kit (TaKaRa, Dalian, China). The quantity and purity of DNA were determined using the spectrophotometric ratio of 260nm/280nm.
DNA sequencingDouble-stranded DNA fragments were generated using ultrasound and converted to blunt ends with T4 DNA polymerase. Klenow Exo was subsequently used to extend the blunt ends. DNA was size-selected in a 2% agarose gel. DNA of 250–250 base pairs was cut and recovered using the gel extraction kit (TaKaRa, Dalian, China) and a DNA library was generated using PCR.
Multiple allele specific PCR (MAS-PCR)MAS-PCR was designed to detect the five most common INH and RIF resistance-conferring mutations, KatG codon 315, mab-inhA: −15, and rpoB codons 516, 526 and 531.11–13 Five pairs primers, two allele-specific primers and three pairs of general detection primers were used simultaneously in a single PCR to produce a PCR band spectrum detecting all mutation variants. The reaction mixture contained the following primers: rpoB516 (1pmol), rpoB526 (5pmol), rpoB531 (32.5pmol), RIRm (30pmol), katgOF (1pmol), katg5R (1pmol), inhAP-15 (6pmol) and inhAPF (6pmol). The other reagents used were 10× PCR buffer (2.5mL), 50mM MgCl2 (2mL), 10mM dNTPs mixture (0.5mL), 5U/mL Taq Polymerase (0.1mL), and DNA template (20ng). The final reaction volume was increased to 25μL by adding dH2O. PCR parameters were 96°C for 3min, 95°C for 50s, 68°C for 40s, 72°C for 60s, repeated for 25 cycles, followed by a 72°C extension for 7min. The PCR product was purified by agarose gel electrophoresis and bands were observed under UV light.
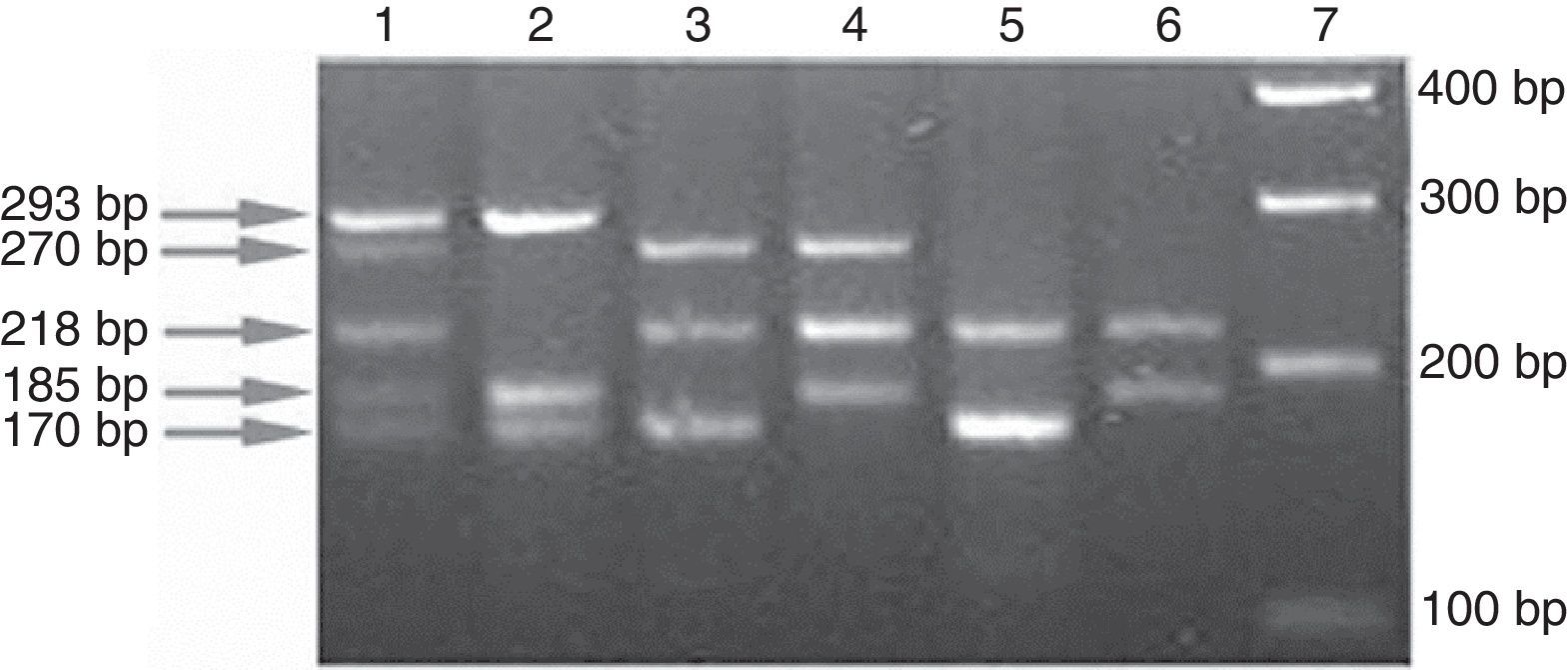
In order to improve sensitivity, the two allele-specific primers were designed such that the point mutation was placed in the second base position from the 3′ end. If the targeted position is the wild-type, the allele specific fragment would be amplified to produce a visible band. In contrast, if the position being targeted contains mutation, it will block the amplification of that specific fragment. An INH and RIF-sensitive isolate will produce five bands (Fig. 1). A mutation in any of the five sites will result in an amplification pattern lacking one or more of these expected bands.
Band patterns indicating drug resistance profile of isolates. Lane 1, H37Rv reference strain (wild-type at all 5 loci); lane 2, mabA-inhA 215CRT and RpoB D516F double mutant; lane 3, KatG S315T and RpoB H526Y double mutant; lane 4, KatG S315G and RpoB 531L double mutant; lane 5, mabA-inhA 215CRT, KatG S315T and RpoB H526D triple mutant; lane 6, mabA-inhA 215CRT, KatG S315T and RpoB S531L triple mutant; and lane 7, DNA ladder.
The DST, DNA sequencing and MAS-PCR results were compared to decide how efficiently each technique can detect INH and RIF tolerance and to compare their sensitivity and specificity. The reproducibility of the MAS-PCR was also assessed.
ResultsDrug sensitivityOf the 191 isolates, 45 were sensitive to INH and RIF, and 103 were resistant to both INH and RIF (Fig. 1). Multidrug resistance detection by DST was as follows: 38/103 resistant to INH and RIF, 36/103 resistant to streptomycin, 27/103 resistant to INH, RIF, streptomycin and ethambutol.
DNA sequencingIn 71/103 (68.9%) isolates, a point mutation was detected in katG codon 315. Of these isolates, 69 isolates harbored the S315T mutation (67 AGCRACC and 2 AGCRACA). In 20/101 (19.8%) isolates, mutation 215CRT on mabA-inhA operon was detected. Of these 20 isolated, four isolates had two point mutations in the operon. RIF resistance mutation is located at the drug resistant determined region (DRDR): S531L mutation was detected in 80/101 (79.2%), H526D/Y mutation was detected in 80/101 (14.9%), and D516F/V mutation was found in 6/101 (5.9%) isolates. No other mutation was found. In short, mutations in these three sites account for 99/101 (98.0%) of RIF resistant isolates. Compared with the gold standard for detecting drug resistance, the sensitivity and specificity of DNA sequencing for detecting INH resistance were 82.9% and 100%, respectively. The sensitivity and specificity of DNA sequencing were 98.0% and 100% for detecting RIF resistance. Because 17 INH resistant isolates lacked the above mentioned mutations in the two sites, DNA sequencing was utilized to detect other potential drug resistance mutation sites. A large deletion was found that extends to the katG gene, removing the last 320bp (e.g., 14.4% of the C-terminus of the protein). Three isolates were found to bear nonsense mutations in codon 198 and 722, which results in a truncated KatG protein lacking the portion of amino acid 740. Moreover, we found a large 500bp repeat (Rv3177–Rv3573) in one isolate.
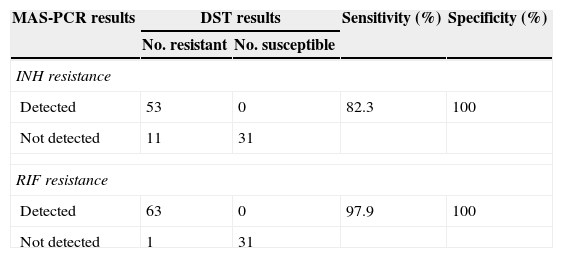
MAS-PCR assayDNA sequencing, DST and MAS-PCR were evaluated for their respective abilities to detect the five target mutation sites. Compared to DNA sequencing, the sensitivity and specificity of MAS-PCR in detecting common mutations were both 100%. In comparison to DST, the sensitivity and specificity of MAS-PCR in detecting common INH resistant mutations were 82.3% and 100%, respectively. The sensitivity and specificity of MAS-PCR in detecting common RIF resistant mutations were 97.9% and 100%, respectively (Table 1). Our results indicate that MAS-PCR has a high degree of reproducibility.
Comparisons of sensitivity and specificity between DST and MAS-PCR in detecting INH and RIF resistance (95 cases).
| MAS-PCR results | DST results | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|
| No. resistant | No. susceptible | |||
| INH resistance | ||||
| Detected | 53 | 0 | 82.3 | 100 |
| Not detected | 11 | 31 | ||
| RIF resistance | ||||
| Detected | 63 | 0 | 97.9 | 100 |
| Not detected | 1 | 31 | ||
Single gene mutations in TB can lead to drug resistance. We designed a MAS-PCR method for the detection of five site mutations, which are related to drug resistance. This method is a fast, and relatively inexpensive test and does not need highly skilled personnel, making it suitable for application in remote areas of China. Use of this technique can reduce the delay in diagnosis and treatment of MDR-TB.
We found that 68.9% of INH resistant isolates contain a KatG S315 T/G mutation and 19.8% contain a 215CRT mutation in the mabA-inhA operon. These two mutations accounted for 82.9% of INH resistant MDR isolates. Our results are consistent with the findings of others, which showed the mutation frequency to be 60.4–93.6% in KatG and 18–50% in 215CRT mabA-inhA.6,14–16 In our region, rpoB codon 531 is the most common mutation site (79.1%). In contrast, mutation frequencies at this locus range from 40.1 to 82.4% in MDR-TB isolates from Brazil, South Africa, China, North Korea, Vietnam, India and Russia.6,14–16 RpoB mutations in codon 516 (5.9%) and 526 (14.9%) accounted for most of the remaining RIF resistant isolates in our study, which is consistent with the reports of mutation frequencies ranging from 2.9 to 16.7% and 5.9 to 40%, respectively. Single RIF resistance is rare and 90% of RIF-resistant M. tuberculosis are also INH resistant strains,17 as was also confirmed in the recently developed Xpert MTB/RIF experiments18 and by our results in this study.
MAS-PCR shows good operability and is 100% accurate as compared with DNA sequencing results in detecting INH and RIF resistance. Moreover, results from the double blind operation using MAS-PCR method in different laboratories are highly consistent. Interestingly, MAS-PCR can detect the H526D mutation portion of RpoB, but not all resistant strains. If MAS-PCR does not detect this particular mutation, this technique can still correctly detect 98.0% of rpoB mutation-containing strains, and 97.1% of RIF-resistant isolates. It is possible that these isolates contain heterologous flora composed with both wild-type and mutation on codon 526 in rpoB gene, resulting in the amplification of the wild-type PCR product. The whole-genome sequencing data also reveals some heterogeneity of these isolates.
DNA sequencing did not reveal any common mutation in 16.4% of INH-resistant and in 1.5% of RIF-resistant isolates. However, a few isolates contain other gene mutations that can allow INH resistance. Three isolates contained mutations that are predicted to produce a truncated KatG protein lacking the C-terminus. The C-terminus of KatG protein has enzymatic function,19,20 and the truncated mutation lacking the C-terminal 41 amino acids is inactivate.20 Although INH resistance is conferred by KatG truncation,21 the minimum protein truncation required to cause INH resistance has not yet been determined. We speculate that the deletion of the C-terminal 19 amino acids is enough to impair the interaction between subunits and leads to INH resistance.
We also found a D94G mutation inside the KatG protein in two isolates. This mutation was found to be unrelated to INH resistance except in one report.22 In this report, the authors found a 34GRA mutation inside the ahpC promoter. In our study, two isolates contained the KatG D94G and AhpC P44R mutations even though AhpC is not obviously associated with the INH-resistance because the overexpression of ahpC in wild-type M. tuberculosis does not increase INH resistance. However, the mutation in ahpC promoter region can be used as a useful marker for detecting INH resistance.8 Whether KatG D94G mutation leads to KatG inactivation or whether AhpC P44R mutation can compensate the inactivation of catalase and peroxidase activity are questions that require further investigation.
In addition, in one isolate we found a 500bp genome repeat crossing Rv3177–Rv3573c. This area contains nat gene (Rv3566c), and this mutated gene confers INH resistance.23 The repeat may result in the upregulation of nat and increase the total activity of the enzyme N-acetyltransferase leading to reduced INH efficacy. However, this needs to be proved experimentally. In five isolates, we did not find any significant mutation in INH resistance related loci. In one of them, no apparent mutation in katG, inhA were found (except in rpoB, where we found a synonymous mutation), suggesting that there may be another DR mechanism.
It should be pointed out that the design of our study was retrospective thus not allowing the estimation of the real incidence of resistance mutations of MDR-TB in the population. Due to the use of previously collected specimens, MAS-PCR were only assessed based on cultured isolates and has yet to be directly assessed using patient's sputum specimens. It is necessary to further assess the effectiveness of MDR-TB using clinical isolates and conduct prospective studies to assess the operability of MAS-PCR relative to DST.
ConclusionGenetic INH and RIF resistance related mutation frequency in the MDR-TB isolates in our area is similar to that in other areas. Due to the high sensitivity and specificity of MAS-PCR in detecting MDR-TB, and the characteristics of this technique including speed, cost-effectiveness and ease of operation, MAS-PCR might be a good method for routine screening of MDR-TB. Further analysis of the genomes of preserved MDR-TB isolates will reveal new mutations associated with INH and RIF resistance.
Conflicts of interestThe authors declare no conflicts of interest.