In order to obtain adequate information for the treatment of methicillin resistant Staphylococcus aureus (MRSA) infections, it is crucial to identify trends in epidemiological and antimicrobial resistance patterns of local S. aureus strains. Community and hospital acquired S. aureus isolates (n=202) were characterized using staphylococcal cassette chromosome mec (SCCmec) typing, pulse field gel electrophoresis (PFGE) analysis, spa typing and minimal inhibitory concentration (MIC) determination. The prevalence of the Panton-Valentine leukocidine (pvl) and several antibiotic resistance genes among the isolates were also detected by PCR. All of the S. aureus isolates were susceptible to vancomycin, daptomycin and linezolid. Three hospital isolates were resistant to teicoplanin while 14 showed intermediate resistance to teicoplanin. The resistance patterns of community-acquired MRSA (CA-MRSA) isolates to other antimicrobials were similar to those of hospital-acquired MRSA (HA-MRSA) isolates except for clindamycin and gentamicin. There was excellent correlation between phenotypes and genotypes in the determination of S. aureus resistance to erythromycin, gentamicin, and tetracycline. The SCCmec type II and SCCmec type IV were the predominant types detected in hospital and community isolates, respectively. The most frequently encountered spa types were t002 and t030 both in HA- and CA-MRSA isolates. Pulsotype A was the most predominant pulsotype identified among the isolates tested, followed by pulsotype B. Seventy-two hospital isolates (19 HA-MRSA and 53 HA-MSSA) and 10 CA-MRSA were positive for the pvl gene. This study shows that the combination of susceptibility testing and various molecular methods has provided useful information on the antibiotic resistance and molecular diversity of S. aureus in a specific region of China. The high proportion of pvl positive MSSA and MRSA isolates observed in this study indicates that adequate measures are needed to curtail the spread of those MRSA and MSSA clones prevailing both in hospital and the community.
Methicillin resistant Staphylococcus aureus (MRSA) is one of the most prevalent nosocomial bacterial pathogens.1–5 causing a wide variety of diseases such as skin and soft tissue infections, bloodstream infections, pneumonia, osteomyelitis and endocarditis, as well as toxin-mediated syndromes like toxic shock and food poisoning.1,4 MRSA was first identified in England in 1961 after which it emerged worldwide.3 MRSA is now responsible for infections with significant morbidity and mortality and it has developed resistance to multiple antimicrobial agents complicating the clinical treatment of infections.4–8 Since the 1990s, MRSA colonization and infections have been increasingly reported among the general population/community and are referred to as community-acquired MRSA (CA-MRSA) strains.2 CA-MRSA strains have been reported in numerous countries such as Australia, New Zealand, the United Kingdom, Canada and the United States.5 In 2008, a study performed in a teaching hospital in Wenzhou, China described and compared the characteristics of hospital and community acquired S. aureus strains.9 Isolates of CA-MRSA are distinct from those of hospital-acquired MRSA (HA-MRSA) in many characteristics which include susceptibility to antimicrobial agents, the type of genetic staphylococcal cassette chromosome mec (SCCmec) elements, and exotoxin gene repertoire.10
Currently, MRSA has become a notorious etiologic agent both in health care facilities and in the community in China, and several studies have shown that this tendency is increasing.2 According to a study published in 2005, the mean incidence of MRSA across China was over 50% with the prevalence in Shanghai being over 80%.11 It is, therefore, of paramount importance to understand the molecular epidemiology and to potentially predict the trends in antibiotic-resistance patterns of MRSA strains both in hospitals and within the community.
It appears that under the selective pressure caused by the extensive use of antibiotics, MRSA clones which carry resistance genes can better adapt to environmental change.6 These resistant genes include the erm genes (ermA, ermB, ermC) encoding erythromycin resistance, the tet genes (tetK, tetL, tetM, tetO) encoding tetracycline resistance and the aac(6′)-aph(2″), aph(3′)-III and ant(4′)-I genes that encode gentamicin resistance.12,13 Resistance determinants can be transposed from one strain to another, sometimes across the species barrier by conjugation, mobilization, phage-mediated transduction, and via as yet unknown mechanisms, resulting in the wide dissemination of antibiotic resistance among circulating staphylococcal strains. Therefore, there is a continuous requirement to determine the effectiveness of these antibiotics against S. aureus infections and the enactment of effective drug policies in China.
The aim of the present study was to investigate the local epidemiological and antimicrobial resistance patterns of S. aureus strains in Shenyang, which is considered necessary to establish empirical and specific therapy guidance for S. aureus treatment. We identified the antibiotic susceptibility patterns and the molecular characteristics of isolates using several methods, including SCCmec typing, pulsed-field gel electrophoresis (PFGE) analysis, spa typing, assessment of several antibiotic resistance genes and minimum inhibitory concentration (MIC) determination. Since MSSA isolates carrying pvl have been reported in the hospital environment and are thought to evolve into MRSA strains through acquisition of a type of SCCmec element that harbors mecA gene,6,14 the prevalence of Panton-Valentine leukocidine (pvl) gene9 among the isolates was also assessed.
Materials and methodsS. aureus isolate collectionThe Shengjing hospital, a 2045-bed tertiary care center located in Shenyang, is one of three affiliated hospitals of the China Medical University. The hospital serves patients mostly coming from three large provinces located in the northeast of China and admits over 70,000 patients annually.
The hospital-acquired S. aureus isolates were obtained from individual patients between January and October 2008 at the Shengjing Hospital of China Medical University in Shenyang from the following departments: internal medicine, nephrology, hematology, gastroenterology, endocrinology, intensive care unit, respiratory medicine, surgical department, surgery intensive care, neurosurgery, thoracic surgery, and pediatrics. In total, 180 hospital-acquired S. aureus were obtained from different clinical specimens, which included sputum (n=94), pus (n=41), blood (n=25), urine (n=9), prostatic fluid (n=4), catheter tips (n=3), drainage fluids (n=3), and ascites (n=1). The definition of hospital-acquired S. aureus infection was as follows: (1) an S. aureus infection identified at least 48h after admission to a hospital; (2) a history of hospitalization, surgery, dialysis, or residence in a long-term care facility within one year of the positive culture date; (3) a permanent indwelling catheter or percutaneous medical device (e.g. tracheostomy tube, gastrostomy tube or Foley catheter) present at the time of culture; or (4) a known positive culture for S. aureus prior to the study period.15 As a comparison, 2103 third-year students from China Medical University (located in the same region as Shengjing hospital) were screened for nasal carriage of S. aureus from whom 22 MRSA were identified. These MRSA strains were deemed to be of community origin since the students were all healthy and health care-related risk factors were absent.2
Only one isolate from each patient or student was used. Isolates were first cultured on sheep blood agar (Becton Dickinson, Cockeysville, MD) at 35°C for 48h. Phenotypic identification of S. aureus was initially based on colony morphology which was then confirmed using the Pos Combo type 41 MicroScan panels (Baxter Diagnostics, Inc., West Sacramento, CA). All the isolates were stored in a storage solution (80% glycerol, 20% brain heart infusion) at −80°C.
Antimicrobial susceptibility testMICs were determined using the broth microdilution method according to the procedure recommended by the Clinical and Laboratory Standards Institute.16,17 The antibiotics tested were penicillin G, oxacillin, tetracycline, erythromycin, and gentamicin (Sigma Chemical Co., St. Louis, MO), cefoxitin (Fujisawa Pharmacy Co., Osaka, Japan), vancomycin (Sigma Chemical, St. Louis, MO), teicoplanin, clindamycin, daptomycin, linezolid, rifampicin, levofloxacin (Daiichi Pharmaceutical Co., Tokyo, Japan), chloramphenicol and trimethoprim-sulfamethoxazole (SXT). The reference strain S. aureus ATCC 29213 was used as a control standard in each test.
DNA extractionGenomic DNA was obtained from 2mL overnight cultures of each isolate using a Biospin Bacteria Genomic DNA Extraction Kit (Biotechs Biological Technology Co., Ltd) and by following the manufacturer's instructions. Five microlitres of lysostaphin (100μg/mL) were added to digest the bacterial cell wall at 37°C for 60min.
PCR detection of the mecA gene and SCCmec typingMethicillin resistant phenotypic identification of the S. aureus isolates was confirmed by the detection of the mecA gene. All isolates were subjected to mecA gene detection by PCR using primers mA1 and mA2 (Table 1). SCCmec typing was performed using PCR according to previous reports.18,19 MRSA strains NCTC10442, N315, 85/2082, CA05 and WIS were used as reference standards for SCCmec typing (I, II, III, IV and V).
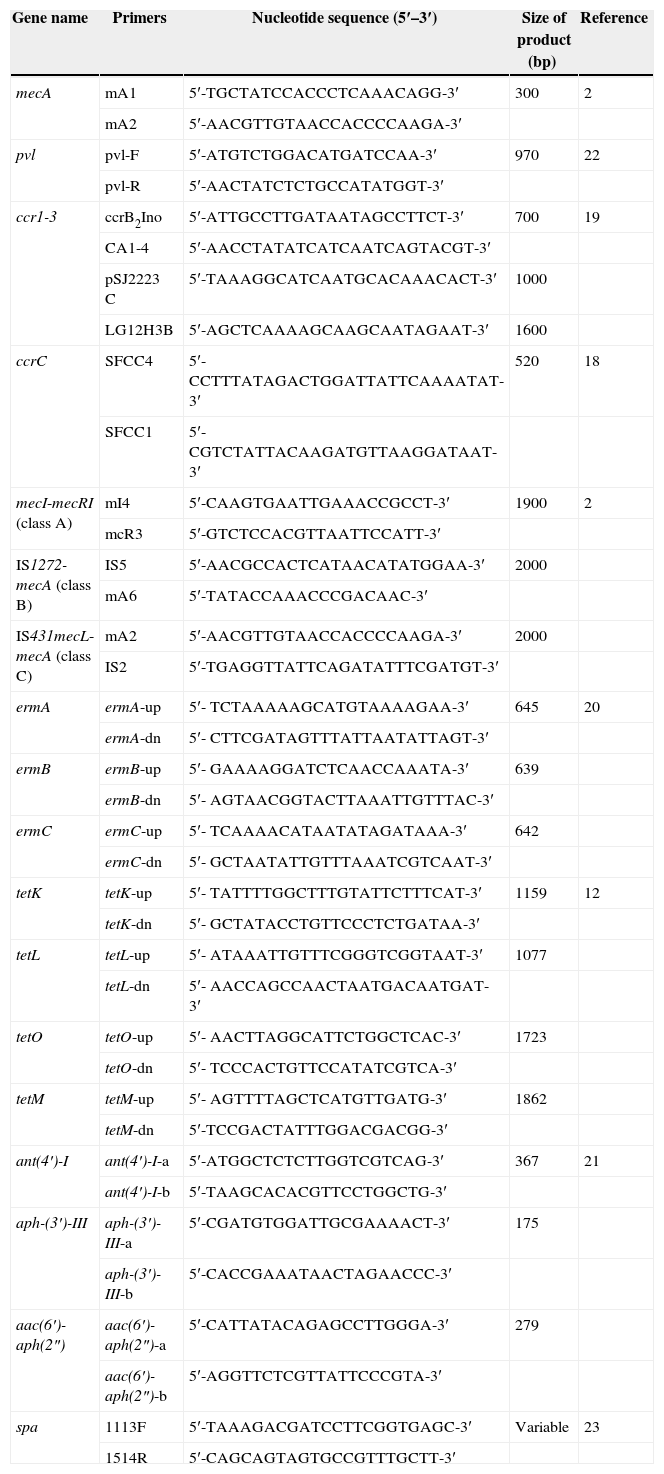
Primers used in this study.
| Gene name | Primers | Nucleotide sequence (5′–3′) | Size of product (bp) | Reference |
|---|---|---|---|---|
| mecA | mA1 | 5′-TGCTATCCACCCTCAAACAGG-3′ | 300 | 2 |
| mA2 | 5′-AACGTTGTAACCACCCCAAGA-3′ | |||
| pvl | pvl-F | 5′-ATGTCTGGACATGATCCAA-3′ | 970 | 22 |
| pvl-R | 5′-AACTATCTCTGCCATATGGT-3′ | |||
| ccr1-3 | ccrB2Ino | 5′-ATTGCCTTGATAATAGCCTTCT-3′ | 700 | 19 |
| CA1-4 | 5′-AACCTATATCATCAATCAGTACGT-3′ | |||
| pSJ2223C | 5′-TAAAGGCATCAATGCACAAACACT-3′ | 1000 | ||
| LG12H3B | 5′-AGCTCAAAAGCAAGCAATAGAAT-3′ | 1600 | ||
| ccrC | SFCC4 | 5′-CCTTTATAGACTGGATTATTCAAAATAT-3′ | 520 | 18 |
| SFCC1 | 5′-CGTCTATTACAAGATGTTAAGGATAAT-3′ | |||
| mecI-mecRI (class A) | mI4 | 5′-CAAGTGAATTGAAACCGCCT-3′ | 1900 | 2 |
| mcR3 | 5′-GTCTCCACGTTAATTCCATT-3′ | |||
| IS1272-mecA (class B) | IS5 | 5′-AACGCCACTCATAACATATGGAA-3′ | 2000 | |
| mA6 | 5′-TATACCAAACCCGACAAC-3′ | |||
| IS431mecL- mecA (class C) | mA2 | 5′-AACGTTGTAACCACCCCAAGA-3′ | 2000 | |
| IS2 | 5′-TGAGGTTATTCAGATATTTCGATGT-3′ | |||
| ermA | ermA-up | 5′- TCTAAAAAGCATGTAAAAGAA-3′ | 645 | 20 |
| ermA-dn | 5′- CTTCGATAGTTTATTAATATTAGT-3′ | |||
| ermB | ermB-up | 5′- GAAAAGGATCTCAACCAAATA-3′ | 639 | |
| ermB-dn | 5′- AGTAACGGTACTTAAATTGTTTAC-3′ | |||
| ermC | ermC-up | 5′- TCAAAACATAATATAGATAAA-3′ | 642 | |
| ermC-dn | 5′- GCTAATATTGTTTAAATCGTCAAT-3′ | |||
| tetK | tetK-up | 5′- TATTTTGGCTTTGTATTCTTTCAT-3′ | 1159 | 12 |
| tetK-dn | 5′- GCTATACCTGTTCCCTCTGATAA-3′ | |||
| tetL | tetL-up | 5′- ATAAATTGTTTCGGGTCGGTAAT-3′ | 1077 | |
| tetL-dn | 5′- AACCAGCCAACTAATGACAATGAT-3′ | |||
| tetO | tetO-up | 5′- AACTTAGGCATTCTGGCTCAC-3′ | 1723 | |
| tetO-dn | 5′- TCCCACTGTTCCATATCGTCA-3′ | |||
| tetM | tetM-up | 5′- AGTTTTAGCTCATGTTGATG-3′ | 1862 | |
| tetM-dn | 5′-TCCGACTATTTGGACGACGG-3′ | |||
| ant(4′)-I | ant(4′)-I-a | 5′-ATGGCTCTCTTGGTCGTCAG-3′ | 367 | 21 |
| ant(4′)-I-b | 5′-TAAGCACACGTTCCTGGCTG-3′ | |||
| aph-(3′)-III | aph-(3′)-III-a | 5′-CGATGTGGATTGCGAAAACT-3′ | 175 | |
| aph-(3′)-III-b | 5′-CACCGAAATAACTAGAACCC-3′ | |||
| aac(6′)-aph(2″) | aac(6′)-aph(2″)-a | 5′-CATTATACAGAGCCTTGGGA-3′ | 279 | |
| aac(6′)-aph(2″)-b | 5′-AGGTTCTCGTTATTCCCGTA-3′ | |||
| spa | 1113F | 5′-TAAAGACGATCCTTCGGTGAGC-3′ | Variable | 23 |
| 1514R | 5′-CAGCAGTAGTGCCGTTTGCTT-3′ |
Antibiotic resistance genes associated with macrolide resistance (ermA, ermB and ermC), aminoglycoside resistance (aac(6′)-aph(2″), aph(3′)-III, ant(4′)-I) and tetracycline resistance (tetK, tetL tetM, tetO) were detected by PCR using previously published protocols and primers as shown in Table 1.12,20,21 All S. aureus isolates were screened for the pvl toxin gene by PCR with primers pvl-1 and pvl-2 as described previously (Table 1).6,22 MRSA strains MW2 was used as a reference standard for pvl gene identification.
spa typingThe spa gene encoding protein A of S. aureus was used for MRSA typing and amplified by PCR with primers 1113F and 1514R (Table 1).23 Sequencing of the spa gene was performed by the Beijing Gene Institution (Beijing, China) using standard methods. The nucleotide sequences were analyzed using the RIDOM Staph-Type software (Ridom GmbH, Germany) to assign a spa type to the various isolates (http://www.ridom.de/spaserver/).
PFGE analysisChromosomal DNAs from the MRSA isolates were digested with SmaI and separated by PFGE using a Gene Path system (CHEF-DR, PULS WAVE.760; Bio-Rad, Hercules, USA) according to the manufacturer's instructions. A 48.5kb ladder was used as a DNA size marker (Bio-Rad, cat. # LS:170-3635). Isolates were considered identical when their PFGE patterns contained the same number and sizes of fragments. PFGE patterns with fewer than 4 band differences between an existing genotype were defined as subtypes of that genotype.2,24 Genotypes were categorized alphabetically.
Statistical analysisStatistical comparisons were applied to determine the significance of differences in resistance rates using Pearson's χ2 test (SPSS version 12.0). p-Values of less than 0.05 were considered statistically significant.
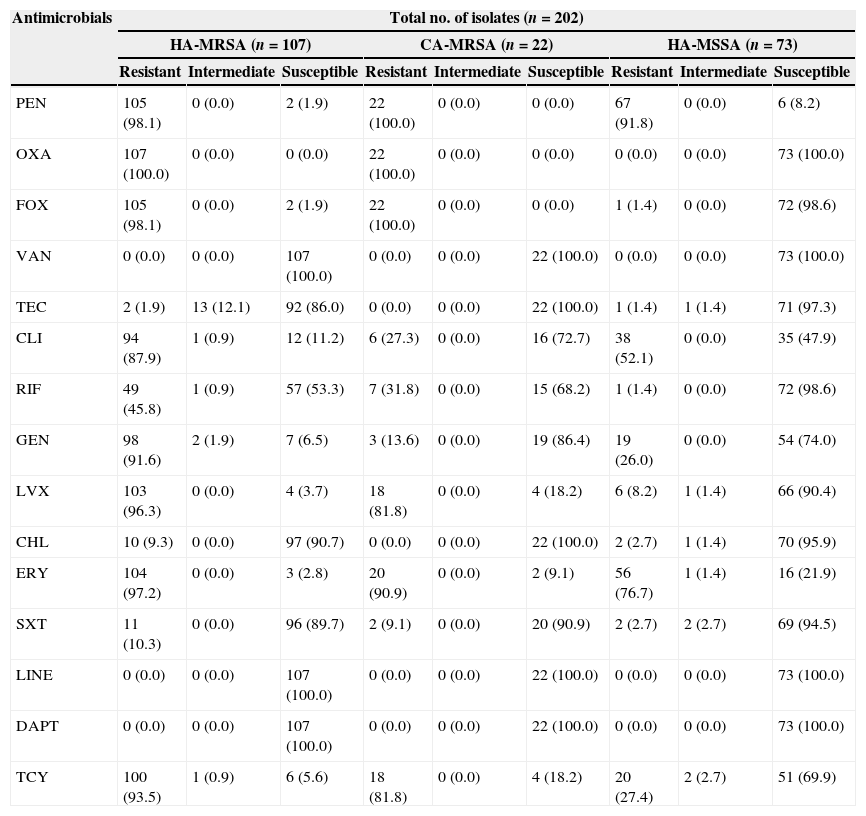
ResultsAntimicrobial susceptibilityOf the 180 hospital-acquired S. aureus isolates identified, 107 were MRSA and 73 were MSSA. Table 2 indicates that all isolates were susceptible to vancomycin, daptomycin, and linezolid. Two HA-MRSA (1.9%) and one HA-MSSA (1.4%) isolates were resistant to teicoplanin and 13 HA-MRSA (12.1%) isolates and one HA-MSSA (1.4%) isolate showed intermediate resistance to teicoplanin. There was no significant difference in the prevalence of teicoplanin resistance between HA-MRSA and HA-MSSA isolates (p>0.05). All of the CA-MRSA isolates were susceptible to vancomycin, daptomycin, linezolid, and teicoplanin. In addition to the antibiotics above, the resistant rates to penicillin G, cefoxitin, erythromycin, levofloxacin, tetracycline, gentamicin, clindamycin were higher for HA-MRSA isolates, at 98.1%, 98.1%, 97.2%, 96.2%, 93.4%, 91.6%, and 87.9%, respectively. In contrast, the resistant rates to rifampicin, chloramphenicol, and SXT were moderately low, at 45.8%, 9.3%, and 10.3%, respectively. The resistance patterns of CA-MRSA isolates to the antibiotics tested were similar to those of the HA-MRSA isolates for most of the antimicrobial agents except for clindamycin and gentamicin.
Antibiotic resistance profile of S. aureus (MSSA and MRSA) from Shenyang, China.
| Antimicrobials | Total no. of isolates (n=202) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HA-MRSA (n=107) | CA-MRSA (n=22) | HA-MSSA (n=73) | |||||||
| Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | |
| PEN | 105 (98.1) | 0 (0.0) | 2 (1.9) | 22 (100.0) | 0 (0.0) | 0 (0.0) | 67 (91.8) | 0 (0.0) | 6 (8.2) |
| OXA | 107 (100.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 73 (100.0) |
| FOX | 105 (98.1) | 0 (0.0) | 2 (1.9) | 22 (100.0) | 0 (0.0) | 0 (0.0) | 1 (1.4) | 0 (0.0) | 72 (98.6) |
| VAN | 0 (0.0) | 0 (0.0) | 107 (100.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 0 (0.0) | 0 (0.0) | 73 (100.0) |
| TEC | 2 (1.9) | 13 (12.1) | 92 (86.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 1 (1.4) | 1 (1.4) | 71 (97.3) |
| CLI | 94 (87.9) | 1 (0.9) | 12 (11.2) | 6 (27.3) | 0 (0.0) | 16 (72.7) | 38 (52.1) | 0 (0.0) | 35 (47.9) |
| RIF | 49 (45.8) | 1 (0.9) | 57 (53.3) | 7 (31.8) | 0 (0.0) | 15 (68.2) | 1 (1.4) | 0 (0.0) | 72 (98.6) |
| GEN | 98 (91.6) | 2 (1.9) | 7 (6.5) | 3 (13.6) | 0 (0.0) | 19 (86.4) | 19 (26.0) | 0 (0.0) | 54 (74.0) |
| LVX | 103 (96.3) | 0 (0.0) | 4 (3.7) | 18 (81.8) | 0 (0.0) | 4 (18.2) | 6 (8.2) | 1 (1.4) | 66 (90.4) |
| CHL | 10 (9.3) | 0 (0.0) | 97 (90.7) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 2 (2.7) | 1 (1.4) | 70 (95.9) |
| ERY | 104 (97.2) | 0 (0.0) | 3 (2.8) | 20 (90.9) | 0 (0.0) | 2 (9.1) | 56 (76.7) | 1 (1.4) | 16 (21.9) |
| SXT | 11 (10.3) | 0 (0.0) | 96 (89.7) | 2 (9.1) | 0 (0.0) | 20 (90.9) | 2 (2.7) | 2 (2.7) | 69 (94.5) |
| LINE | 0 (0.0) | 0 (0.0) | 107 (100.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 0 (0.0) | 0 (0.0) | 73 (100.0) |
| DAPT | 0 (0.0) | 0 (0.0) | 107 (100.0) | 0 (0.0) | 0 (0.0) | 22 (100.0) | 0 (0.0) | 0 (0.0) | 73 (100.0) |
| TCY | 100 (93.5) | 1 (0.9) | 6 (5.6) | 18 (81.8) | 0 (0.0) | 4 (18.2) | 20 (27.4) | 2 (2.7) | 51 (69.9) |
PEN, penicillin G; OXA, oxacillin; FOX, cefoxitin; VAN, vancomycin; TEC, teicoplanin; CLI, clindamycin; RIF, rifampicin; GEN, gentamicin; LVX, levofloxacin; CHL, chloramphenicol; ERY, erythromycin; SXT, trimethoprim-sulfamethoxazole; TCY, tetracycline; LINE, linezolid; DAPT; daptomycin.
Compared to HA-MRSA isolates, the HA-MSSA isolates presented lower resistant rates to almost all antibiotics tested. HA-MSSA isolates showed resistance only to penicillin G, erythromycin, clindamycin, tetracycline and gentamicin to some degree. The predominant antibiotypes among the HA-MRSA isolates were resistant to penicillin G, oxacillin, cefoxitin, erythromycin, tetracycline, gentamicin, levofloxacin, and clindamycin. The predominant antibiotypes among the HA-MSSA isolates were those showing resistance to penicillin G, erythromycin, and clindamycin.
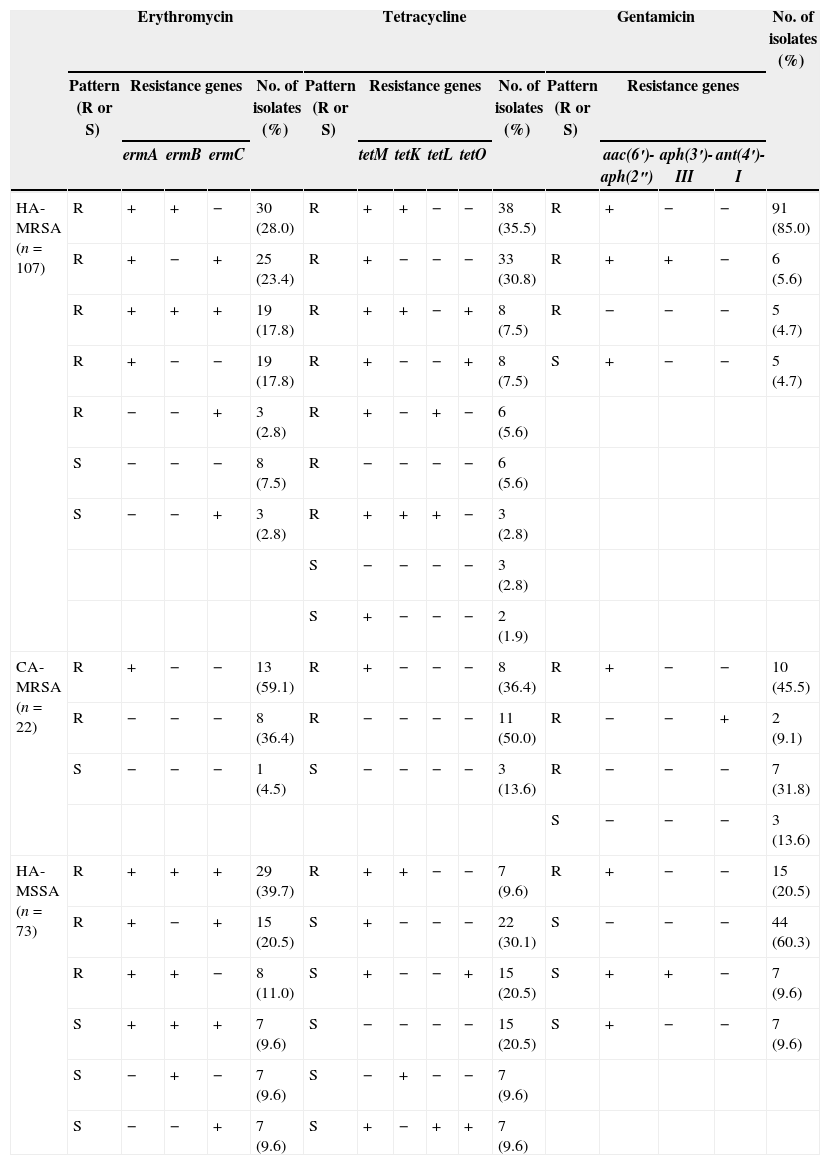
Antimicrobial resistance genesTable 3 shows that the ermA gene was frequently detected in both HA-MRSA (93/107, 86.9%) and HA-MSSA (59/73, 80.8%) isolates while the detection rates of the ermB and ermC genes were significantly higher in HA-MSSA isolates (51/73, 69.9%; 58/73, 79.5%) compared to HA-MRSA isolates (49/107, 45.8%; 50/107, 46.7%). The triple combination of ermA, ermB and ermC was present in 17.8% of HA-MRSA isolates and 49.3% of HA-MSSA isolates. In contrast, the CA-MRSA isolates only possessed the ermA gene, which was detected in 13 (59.1%) erythromycin-resistant isolates.
Characterization of hospital isolates from Shenyang based on antibiotic susceptibility patterns and detection of antibiotic resistance genes.
| Erythromycin | Tetracycline | Gentamicin | No. of isolates (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pattern (R or S) | Resistance genes | No. of isolates (%) | Pattern (R or S) | Resistance genes | No. of isolates (%) | Pattern (R or S) | Resistance genes | |||||||||
| ermA | ermB | ermC | tetM | tetK | tetL | tetO | aac(6′)-aph(2″) | aph(3′)-III | ant(4′)-I | |||||||
| HA-MRSA (n=107) | R | + | + | − | 30 (28.0) | R | + | + | − | − | 38 (35.5) | R | + | − | − | 91 (85.0) |
| R | + | − | + | 25 (23.4) | R | + | − | − | − | 33 (30.8) | R | + | + | − | 6 (5.6) | |
| R | + | + | + | 19 (17.8) | R | + | + | − | + | 8 (7.5) | R | − | − | − | 5 (4.7) | |
| R | + | − | − | 19 (17.8) | R | + | − | − | + | 8 (7.5) | S | + | − | − | 5 (4.7) | |
| R | − | − | + | 3 (2.8) | R | + | − | + | − | 6 (5.6) | ||||||
| S | − | − | − | 8 (7.5) | R | − | − | − | − | 6 (5.6) | ||||||
| S | − | − | + | 3 (2.8) | R | + | + | + | − | 3 (2.8) | ||||||
| S | − | − | − | − | 3 (2.8) | |||||||||||
| S | + | − | − | − | 2 (1.9) | |||||||||||
| CA-MRSA (n=22) | R | + | − | − | 13 (59.1) | R | + | − | − | − | 8 (36.4) | R | + | − | − | 10 (45.5) |
| R | − | − | − | 8 (36.4) | R | − | − | − | − | 11 (50.0) | R | − | − | + | 2 (9.1) | |
| S | − | − | − | 1 (4.5) | S | − | − | − | − | 3 (13.6) | R | − | − | − | 7 (31.8) | |
| S | − | − | − | 3 (13.6) | ||||||||||||
| HA-MSSA (n=73) | R | + | + | + | 29 (39.7) | R | + | + | − | − | 7 (9.6) | R | + | − | − | 15 (20.5) |
| R | + | − | + | 15 (20.5) | S | + | − | − | − | 22 (30.1) | S | − | − | − | 44 (60.3) | |
| R | + | + | − | 8 (11.0) | S | + | − | − | + | 15 (20.5) | S | + | + | − | 7 (9.6) | |
| S | + | + | + | 7 (9.6) | S | − | − | − | − | 15 (20.5) | S | + | − | − | 7 (9.6) | |
| S | − | + | − | 7 (9.6) | S | − | + | − | − | 7 (9.6) | ||||||
| S | − | − | + | 7 (9.6) | S | + | − | + | + | 7 (9.6) | ||||||
In this study, the HA-MRSA isolates were more likely to contain the tetM (91.6% versus 69.9%), tetK (45.8% versus 19.2%) and tetL (11.2% versus 9.6%) genes and less likely to contain tetO (15.0% versus 30.1%) than the HA-MSSA isolates. Among the CA-MRSA isolates, 86.4% (19/22) were resistant to tetracycline. Only eight of these isolates possessed the tetM gene.
The aminoglycoside-resistance gene aac(6′)-aph(2″) was identified most frequently in HA-MRSA isolates (102/107, 95.3%). The aph(3′)-III gene was detected in six (5.6%) HA-MRSA and in seven (9.6%) HA-MSSA isolates but was not detected in any of the CA-MRSA isolates. Interestingly, the ant(4′)-I gene was detected in two CA-MRSA isolates (9.1%) but was not detected in any of the hospital isolates.
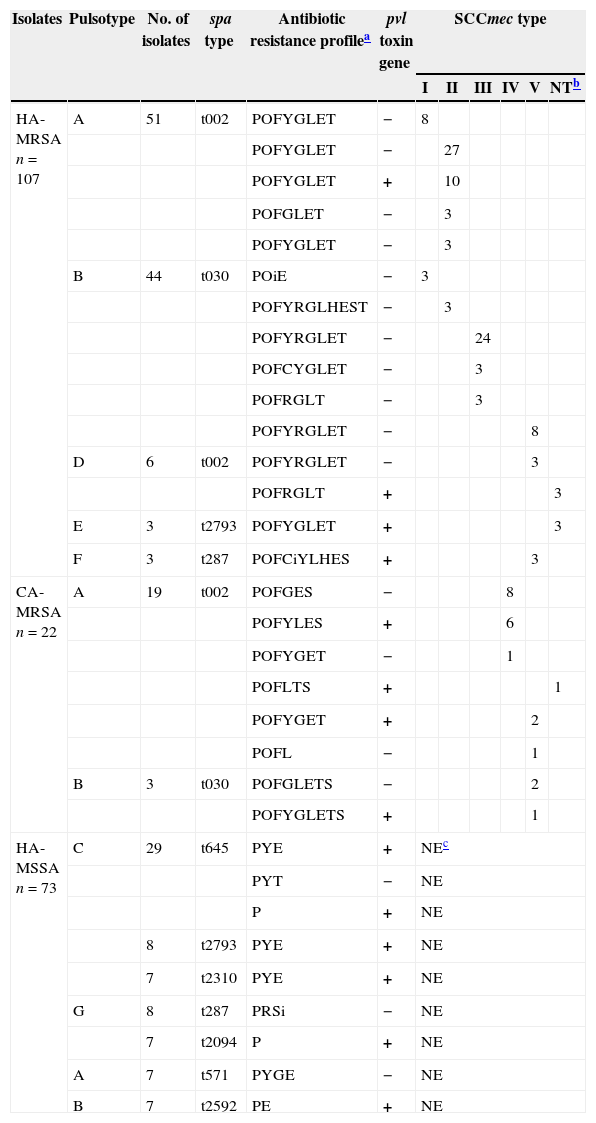
Molecular characteristics of CA-MRSA and HA-S. aureus isolatesSCCmec types were identified in 107 HA-MRSA and 22 CA-MRSA isolates as shown in Table 4. Six HA-MRSA and one CA-MRSA isolate(s) could not be assigned SCCmec types using the established protocols. For the HA-MRSA isolates, the most frequently detected SCCmec types were SCCmec type II (43.0%, 46/107) and SCCmec type III (28.0%, 30/107), followed by SCCmec type V (13.1%, 14/107) and SCCmec type I (10.3%, 11/107). For the CA-MRSA, SCCmec type IV (68.2%; 15/22) and V (27.3%; 6/22) were the most frequently found.
Molecular characteristics of isolates from hospital and community settings and detection of virulence genes.
| Isolates | Pulsotype | No. of isolates | spa type | Antibiotic resistance profilea | pvl toxin gene | SCCmec type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | NTb | ||||||
| HA-MRSA n=107 | A | 51 | t002 | POFYGLET | − | 8 | |||||
| POFYGLET | − | 27 | |||||||||
| POFYGLET | + | 10 | |||||||||
| POFGLET | − | 3 | |||||||||
| POFYGLET | − | 3 | |||||||||
| B | 44 | t030 | POiE | − | 3 | ||||||
| POFYRGLHEST | − | 3 | |||||||||
| POFYRGLET | − | 24 | |||||||||
| POFCYGLET | − | 3 | |||||||||
| POFRGLT | − | 3 | |||||||||
| POFYRGLET | − | 8 | |||||||||
| D | 6 | t002 | POFYRGLET | − | 3 | ||||||
| POFRGLT | + | 3 | |||||||||
| E | 3 | t2793 | POFYGLET | + | 3 | ||||||
| F | 3 | t287 | POFCiYLHES | + | 3 | ||||||
| CA-MRSA n=22 | A | 19 | t002 | POFGES | − | 8 | |||||
| POFYLES | + | 6 | |||||||||
| POFYGET | − | 1 | |||||||||
| POFLTS | + | 1 | |||||||||
| POFYGET | + | 2 | |||||||||
| POFL | − | 1 | |||||||||
| B | 3 | t030 | POFGLETS | − | 2 | ||||||
| POFYGLETS | + | 1 | |||||||||
| HA-MSSA n=73 | C | 29 | t645 | PYE | + | NEc | |||||
| PYT | − | NE | |||||||||
| P | + | NE | |||||||||
| 8 | t2793 | PYE | + | NE | |||||||
| 7 | t2310 | PYE | + | NE | |||||||
| G | 8 | t287 | PRSi | − | NE | ||||||
| 7 | t2094 | P | + | NE | |||||||
| A | 7 | t571 | PYGE | − | NE | ||||||
| B | 7 | t2592 | PE | + | NE | ||||||
The spa types were assigned by sequencing analysis of PCR products of the spa gene (Table 4). Nine spa types (t002, t030, t645, t2793, t287, t2310, t2094, t571 and t2592) were found in 180 of the hospital-acquired isolates and two (t002 and t030) were found in 22 of the CA-MRSA isolates. The most frequently encountered spa types were shown to be t002 and t030 both in HA-MRSA (n=57; n=44) and CA-MRSA isolates (n=19; n=3).
A total of seven pulsotypes were identified among the isolates and they were classified into seven major groups (A to G) according to their band patterns (data not shown). As shown in Table 4, the majority of the S. aureus isolates (n=77), including both hospital- and community-acquired isolates, belonged to the pulsotype A cluster, followed by pulsotypes B, C and G.
Prevalence of the pvl geneOf 202 S. aureus isolates tested, 72 HA isolates (e.g., 19 HA-MRSA and 53 HA-MSSA) and 10 CA-MRSA isolates were positive for the pvl gene. All of the 53 HA-MSSA isolates positive for pvl were obtained from pus while 19 of the pvl positive HA-MRSA isolates were obtained from 13 sputum and 6 ascite samples. Further statistical analysis showed that patients with pvl positive HA-MRSA isolates were older than pvl positive HA-MSSA carriers (65.8 year versus 11.9 year; p<0.05). Two SCCmec types were identified among the 19 pvl positive HA-MRSA isolates, namely the SCCmec type II (10/107; 9.3%) and type V (3/107; 2.8%). Among the 22 CA-MRSA isolates, the pvl positive isolates were SCCmec type IV (6/22; 27.3%) and type V (3/22; 13.6%).
DiscussionThis study reveals that the HA-MRSA, HA-MSSA and CA-MRSA isolates differ significantly in antimicrobial susceptibility (Table 2). Considering the low resistance rates of HA-MRSA and CA-MRSA to vancomycin, teicoplanin, chloramphenicol, daptomycin, linezolid and SXT, these antimicrobials may still be effective in therapy for S. aureus infections. In contrast, the high resistance rates of isolates to penicillin, oxacillin, cefoxitin, levofloxacin, erythromycin, tetracycline, strongly indicates that the use of these antibiotics should not be encouraged for treating patients with such S. aureus infections in the Shenyang region of China.
All S. aureus isolates were susceptible to vancomycin, daptomycin and linezolid. However two (1.9%) HA-MRSA and one (1.4%) HA-MSSA isolates were resistant to teicoplanin while 13 (12.1%) HA-MRSA and one (1.4%) HA-MSSA isolate showed intermediate resistance to this antibiotic. Glycopeptides including vancomycin and teicoplanin are of great importance in the treatment of patients with penicillinase-producing and methicillin-resistant S. epidermidis and S. aureus infections.25 However, along with the increasing use of vancomycin and teicoplanin in clinical practice, there have been numerous reports on the emergence of MRSA with reduced susceptibility to glycopeptides in addition to teicoplanin therapy failure.26–28 Since our study showed that teicoplanin resistance was present in HA-MRSA and HA-MSSA isolates, it would seem prudent to routinely screen both MRSA and MSSA isolates for reduced susceptibility to both vancomycin and teicoplanin. Our results also indicated that daptomycin and linezolid remain effective in treating those MRSA or MSSA strains exhibiting teicoplanin intermediate resistance.
As shown in Table 2, about 97.2%, 93.5%, 91.6% and 87.9% of the HA-MRSA isolates were resistant to erythromycin, tetracycline, gentamicin and clindamycin, respectively. In addition, 13.6% and 27.3% of CA-MRSA isolates were resistant to gentamicin and clindamycin. Compared to the HA-MRSA isolates, the CA-MRSA isolates were therefore more likely to be susceptible to gentamicin and clindamycin. Clindamycin is recommended by current guidelines as an option for the management of CA-MRSA infections.29 Nevertheless, resistance to clindamycin was detected in 27.3% of the CA-MRSA in this study, a fact that should be taken into consideration in the future management of CA-MRSA. Most of the HA-MSSA and CA-MRSA isolates manifested susceptibility to several antimicrobials excluding the β-lactams (e.g. penicillin G, oxacillin, cefoxitin), and both CA-MRSA and HA-MSSA isolates showed low resistance to gentamicin. However, compared to CA-MRSA, HA-MSSA isolates were more likely to have high-level resistance to clindamycin. It may be that the HA-MSSA isolates demonstrate reduced susceptibility to clindamycin due to the selective pressures in the clinical setting.
Table 3 demonstrates that ermA was the most common gene encoding macrolides resistance in both the hospital and community acquired S. aureus isolates. The ermB and ermC genes were more prevalent in HA-MSSA than in HA-MRSA isolates. Other investigators have also found that ermA is the dominant erm gene in S. aureus.13Table 3 shows that three HA-MRSA and 21 HA-MSSA isolates were susceptible to erythromycin even though they possessed at least one of the three erm genes. Similarly, Sekiguchi et al. 30 found discordance among phenotypic erythromycin susceptibility and the presence of erm genes. They stated that this discordance might be attributed to a mutation in the coding or promoter region of the genes. We also noted that eight of CA-MRSA were resistant to erythromycin but did not possess any erythromycin resistance genes as determined by PCR (Table 3). This might be explained by the location of these genes on small plasmids, which were occasionally lost.20,31,32 The most frequently identified gene implicated in aminoglycoside resistance in our study was aac(6′)-aph(2″), which was found in 95.3% of HA-MRSA, 45.5% of CA-MRSA and 39.7% of HA-MSSA isolates, respectively. The Aph(3′)-III gene, infrequently found in S. aureus, was identified in 5.6% of HA-MRSA and 9.6% of HA-MSSA isolates, respectively. The ant(4′)-I gene, another infrequently found gene in S. aureus, was detected in 9.1% of CA-MRSA isolates.
The use of several different molecular methods is currently necessary for the accurate typing of MRSA, both in outbreak situations and for international comparisons.33 In this study, PFGE analysis indicated that the HA-MRSA isolates could be grouped into five different types (Pulsotype A, B, D, E and F), pulsotype A was the most predominant type (51/107; 47.7%), followed by pulsotype B (44/107; 41.1%). Concurrently, CA-MRSA isolates were grouped into two different types (Pulsotype A and B), 19 belonged to pulsotype A and three strains belonged to pulsotype B. The spa typing revealed four spa types including t002, t030, t2793 and t287 among the HA-MRSA isolates, while two spa types of t002 and t030 were identified in CA-MRSA. The results demonstrated that the predominant spa types (t002 and t030) and pulsotypes (A and B) in community acquired MRSA were also predominant among hospital isolates, which suggested that they shared the same clonal origin. This also indicates that the cross-transmission of S. aureus strains between the community and hospital environment may have occurred which is in accordance with previous reports documenting increasing colonization and infection due to MRSA strains of community origin in hospitals and vice versa.34,35
SCCmec typing of HA-MRSA isolates revealed that 13.1% (14/107) strains carried SCCmec type V, which is mainly carried by CA-MRSA isolates36 indicating that HA-MRSA and CA-MRSA isolates might not be distinguished merely by identifying their SCCmec genotypes. Ma et al.10 reported that 49 of 51 strains (96.0%) isolated from both community- and healthcare-associated cases in Uruguay carried either SCCmec type IV or SCCmec type V elements. In recent years, there have been numerous reports describing nosocomial outbreaks caused by strains harboring SCCmec type IV, which was originally identified in CA-MRSA strains.37,38 Compared with SCCmec type I, II and III, SCCmec type IV and type V are small in size and are more prone to transfer between staphylococcal species, presumably via phage-associated transduction.2,18,36 In this study, seven MRSA strains could not be assigned SCCmec types using the established protocols. Presently work is being undertaken to assign SCCmec types to those seven strains using recently reported primers.45
Worldwide pandemic MRSA clones have been reported as the New York/Japan clone (ST5-type II SCCmec), the pediatric clone (ST5-type IV SCCmec), the archaic clone (ST250-type I SCCmec), the Iberian clone (ST247-SCCmec type 1A), and the Brazilian/Hungarian clone (ST239-type III SCCmec) by Oliveira et al. 39 In the present study, we identified several different MRSA clones that were distinguishable based on SCCmec typing, spa typing, and distinct antimicrobial resistance profiles (Table 4). We conclude that spa types t030 and t002 were the two most common spa types identified in this study. Most of the t030 belong to the Brazilian ST239-MRSA-III epidemic clones while the t002 belong to both the New York/Japan ST5-MRSA-II and the pediatric ST5-MRSA-IV epidemic clones which are the most prevalent in Asian countries, especially China.11,40
It is known that the pvl gene locus is carried on bacteriophages, namely φPVL, φSLT, φSa2mw and φ108PVL, which encode pvl components. S. aureus isolates producing PVL have been strongly associated with severe skin and soft-tissue infections (SSTIs), and occasionally, severe necrotizing pneumonia.6 Some previous studies have described a low prevalence of pvl genes (<5%) in HA-MRSA,41,42 while pvl positive CA-MRSA clones have spread globally.2,43 In our study, the pvl gene was identified in 17.8% (19/107) of HA-MRSA and 45.5% (10/22) CA-MRSA isolates, respectively. In Taiwan, during 2005 the prevalence of pvl positive MRSA isolates was 23.3% in hospitals.44 Yu et al. also reported the prevalence of the pvl gene among clinical isolates of S. aureus in a teaching hospital, and most of the pvl positive isolates were SCCmec type III.9 Furthermore, in this study we also found a high prevalence of pvl positive MSSA isolates of hospital origin.
Overall, the present study suggests a bi-directional movement of S. aureus strains between healthcare and community settings in Shenyang, China. The spread of pvl positive MRSA clones with specific genotypes within hospitals and the community is a particular cause of concern and could have important implications for the implementation of effective infection control strategies and measures.
ConclusionsThis study has shown that the combination of susceptibility testing and various molecular methods provides useful information on the antibiotic resistance and molecular diversity of S. aureus in a region of Northeastern China. Although the number of S. aureus isolates available for our investigation together with relevant epidemiological data was limited, the high proportion of pvl positive MRSA and MSSA isolates observed in this study indicates that adequate measures are needed in order to curtail and control the spread and establishment of such MRSA and MSSA isolates in Chinese health care institutions.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank professors Keiiti Hiramatsu and Teruyo Ito (Juntendo University) for providing MRSA strains as SCCmec typing standards. This work was supported by the National Natural Science Foundation (No. 30972520) and the Ministry of Education in China (No. [2009] 1001).