Carbapenem resistance in members of order Enterobacterales is a growing public health problem causing high mortality in developing and industrialized countries. Its emergence and rapid propagation worldwide was due to both intercontinental spread of pandemic strains and horizontal dissemination via mobile genetic elements (MGE) such as plasmids and transposons.
ObjectiveTo describe MGE carrying carbapenem resistance genes in Enterobacterales which have been reported in South America.
Search strategy and selection criteriaA search of the literature in English or Spanish published until 2019 in PubMed, Google Scholar, LILACS and SciELO databases was performed for studies of MGE in Enterobacterales reported in South American countries.
ResultsSeven South American countries reported MGE related to carbapenemases. Carbapenemase-producing Klebsiella pneumoniae belonging to clonal complex 258 were the most prevalent pathogens reported; others carbapenemase-producing Enterobacterales such as Escherichia coli, Serratia marcescens, and Providencia rettgeri also have been reported. The MGE implicated in the spread of the most prevalent carbapenemase genes are Tn4401 and non-Tn4401 elements for blaKPC and ISAba125 for blaNDM, located in different plasmid incompatibility groups, i.e. L/M, A/C, FII and bacterial clones.
ConclusionThis review indicates that, like in other parts of the world, the most commonly reported carbapenemases in Enterobacterales from South America are being disseminated through clones, plasmids, and transposons which have been previously reported in other parts of the world.
Enterobacterales are the most prevalent opportunistic pathogens in hospital settings, causing sepsis, pneumonia, soft tissue infections, urinary tract infections, and others. To make this problem worse, the emergence of resistance to β-lactams, the most commonly used family of antibiotics, has drastically reduced treatment options for those infections.1 Since Alexander Fleming described the penicillin in 1928, a large number of β-lactams have been developed belonging to five groups: penams, penems, cephems, monobactams, and carbapenems, all of which possess a β-lactam ring. The carbapenems group is the election to treat infections caused by bacteria producing extended-spectrum β-lactamases (ESBLs) or AmpC cephalosporinases.2
In recent years, the carbapenem-resistant Enterobacterales (CRE) have rapidly emerged and disseminated worldwide. This resistance is caused by either production of carbapenemases or a combination of mechanisms such as the presence of different ESBLs (or AmpC) in addition to porin loss. The evidence suggests that clonal dissemination of carbapenemase-producing Enterobacterales (CPE) plays a critical role in hospital outbreaks of life-threatening infections, which have increased mortality, morbidity and hospitalization costs.3 However, mobile genetic elements (MGEs) including plasmids, transposable elements (TEs), and integrons are probably the most important factors implicated with the dissemination of carbapenemase genes among different bacterial species. TE, including insertion sequences (ISs) and transposons (Tns), are discrete DNA structures, located in plasmids or chromosomes, which can move (through the activity of self-encoded transposases) to new locations in the genome. The ISs are the smallest TEs that can move resistance genes as part of a composite transposon; a gene bounded by two copies of the same or related IS that can move as a single unit.4,5 In this review, we describe different MGEs associated with carbapenemase genes in Enterobacterales members reported in South American countries.
MethodologyA search of literature published in English or Spanish until August 2019 was performed using PubMed, Google Scholar, LILACS and SciELO databases. In a first-round, the keywords ‘mobile genetic elements AND carbapenemase resistance, carbapenemase AND Enterobacteriaceae or Enterobacterales AND (Argentina OR Brazil OR Bolivia OR Colombia OR Chile OR Ecuador OR Guyana OR French Guiana OR Paraguay OR Peru OR Suriname OR Uruguay OR Venezuela)’ were used. Abstracts or conference reports were not included unless the article form was unavailable. All selected articles were subjected to a second search round for selection of specific studies using molecular analysis criteria: (a) description of clonality based in sequence type (ST) using multilocus sequence typing (MLST) or whole genome sequencing and (b) descriptions of MGEs associated with carbapenemase genes in Enterobacterales species, including plasmid incompatibility groups (based on replicon typing or restriction digestion performed with S1 nuclease) and/or description of TE or integrons structures using DNA sequencing.
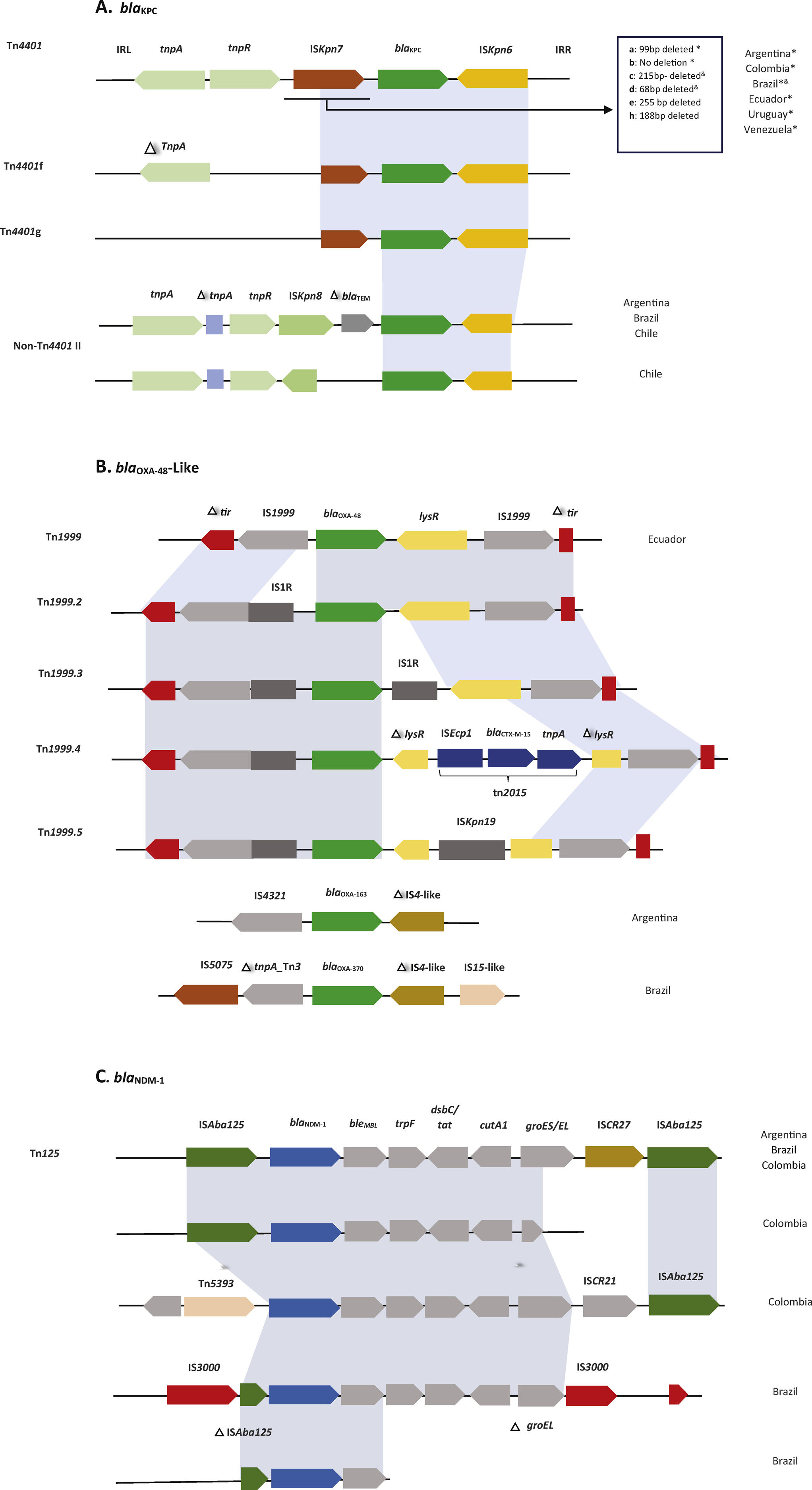
ResultsArgentinaThe first report of chromosomally encoded NMC-A carbapenemase was in an Enterobacter cloacae isolate recovered from a leukemia patient in 2004.6 Four years later a KPC-2-producing K. pneumoniae strain was reported.7 Gomez et al. described the Tn4401a isoform associated with blaKPC-2 harbored in the international clone K. pneumoniae ST258. Interestingly, a blaKPC-2 gene was associated with non-Tn4401 elements (NTE) in the non-ST258 clones of K. pneumoniae (ST11, ST476, and ST526). In other Enterobacteriaceae species, it was harbored in transferable plasmids belonging to IncHI2, IncL/M and IncA/C incompatibility groups.8 TheblaNDM-1 gene was located in a Tn125 composite transposon harbored in plasmids of different sizes, reported in Providencia rettgeri.9 A blaIMP-8-harboring plasmid IncA/C1-ST13 was described in E. coli, and the carbapenemase gene was associated with integron class I flanked by two IS26 elements.10 Other metallo-β-lactamases (MBL), such as blaVIM-2 and blaVIM-11, were associated with integrons In883, In885, In346, In900 in E. cloacae,11 and blaVIM-16 associated with class I integron in Serratia marcescens.12 Two genetically related K. pneumoniae isolates were recovered from the same patient, one isolate harboring blaOXA-163 and the other blaOXA-247. Interestingly, their genetic environment in both genes comprised the insertion sequence IS4321 upstream and IS4-like insertion downstream.13
ColombiaThe first description of KPC-2-producing K. pneumoniae isolate in South America was reported in Colombia in 2005.14 Later, a KPC-3-producing K. pneumoniae was isolated from an Israeli patient (international spread).15 Another report showed that the blaKPC-2 was more prevalent than blaKPC-3 in ST258 and non-ST258K. pneumoniae isolates, and the isoform Tn4401a was more prevalent than Tn4401b.16 Other reports showed a close relationship of blaKPC-2 with Tn4401b isoform (harbored in IncL/M plasmids) in isolates of K. pneumoniae ST14, ST338, and ST339.17 Interestingly, the isoform “b” was also harbored in IncA/C and IncF plasmids in non-K. pneumoniae isolates.18blaNDM-1 was reported in Escherichia coli associated with Tn125 and Tn5393, located in IncA/C plasmids.19 An outbreak caused by a K. pneumoniae ST1043 carrying an IncA/C, blaNDM-1-plasmid was also reported.20
BrazilCarbapenem-resistant K.pneumoniae is the most common CPE pathogen in this country. KPC-2-producing K. pneumoniae belonging to the clonal complex (CC) 258 (including ST258, ST11, and ST437) has been described in a variety of MGEs associations (IncFII, IncN, IncL/M, and untypable plasmids carrying Tn4401a or Tn4401b), which were successfully disseminated among species of Enterobacterales.21 Similarly, another study described a KPC-2-producing K. pneumoniae ST11 carrying a Tn4401 located in a IncW plasmid group.22 Recent work described two K. pneumoniae isolates carrying the blaKPC-2 gene in NTE IId located in IncQ1 and Col-like plasmids.23 In one study the authors showed an association of blaKPC-2 with Tn4401b and IncN plasmids in CC11K. pneumoniae.24E. coli, E. cloacae, Enterobacter aerogenes, Citrobacter freundii have also been reported carrying blaKPC-2 gene located in Tn4401b or Tn4401d isoforms on plasmids of different sizes.25 A new carbapenemase, blaBKC-1, was reported in K. pneumoniae ST1781 and it was classified as a new member of molecular Ambler class A serine carbapenemases in 2015; it was located in a 10-kb non-conjugative IncQ plasmid that included a mobilization system, ISKpn23.26
Reports of MBL and their MGEs were related to blaNDM-1 and associated with a truncated ISAba125 in Enterobacterhormaechei27 or inside a ∼10kb, composite transposon (two copies of ISAba125) named Tn125 in P. rettgeri.28 Interestingly, this gene has also been located in the transposon Tn3000 in E. coli and E. hormaechei.29 Other MBLs such blaIMP-1 gene in K. pneumoniae and P.rettgerii30,31 and blaIMP-10 in S.marcescens32 were linked to class 1 integrons.
The blaOXA-370 reported in E. hormaechei differed from OXA-48 only by a single amino acid substitution. This gene was flanked upstream by a Tn3 family transposase gene tnpA (truncated by an IS5075-like insertion) and downstream of a Tn4 family tnpA gene (truncated by an IS15-like insertion).33
EcuadorA blaKPC-2 gene was detected in K. pneumoniae ST258 and ST25 and associated with Tn4401a (Reyes et al., manuscript in preparation), whereas the blaNDM-1 gene harbored in IncA/C plasmid was reported in K. pneumoniae ST147.34 A blaOXA48-like gene harbored in a Tn1999 was reported in a clinical isolate Raoultella ornithinolytica.35
ChileThe Tn4401a isoform harboring blaKPC gene was reported in K. pneumoniae ST258, ST101, ST25 clones and an NTE variant 1 in K. pneumoniae ST11, ST1161, and ST29.36
UruguayThe presence of blaKPC-2 gene harbored in Tn4401a was identified in K. pneumoniae ST 258 causing an outbreak.37
VenezuelaThe blaKPC-2 located in a Tn4401b was reported in K. pneumoniae ST11, ST15, ST833, ST1271, ST1857, ST1859 and ST1860 clones.38K. pneumoniae ST833 was also described carrying blaKPC-2 and a class 1 integron harboring blaVIM-2.39
DiscussionAlthough some successful bacterial clones can disseminate resistance vertically (e.g. K. pneumoniae belonging to CC258 carrying Tn4401/blaKPC), carbapenemase genes (as well as other antibiotic resistance determinants) are frequently transmitted horizontally between different enterobacterial clones, species, and genera.40 Furthermore, these genes also disseminate among different plasmids and chromosomes through transposition or recombination events.41
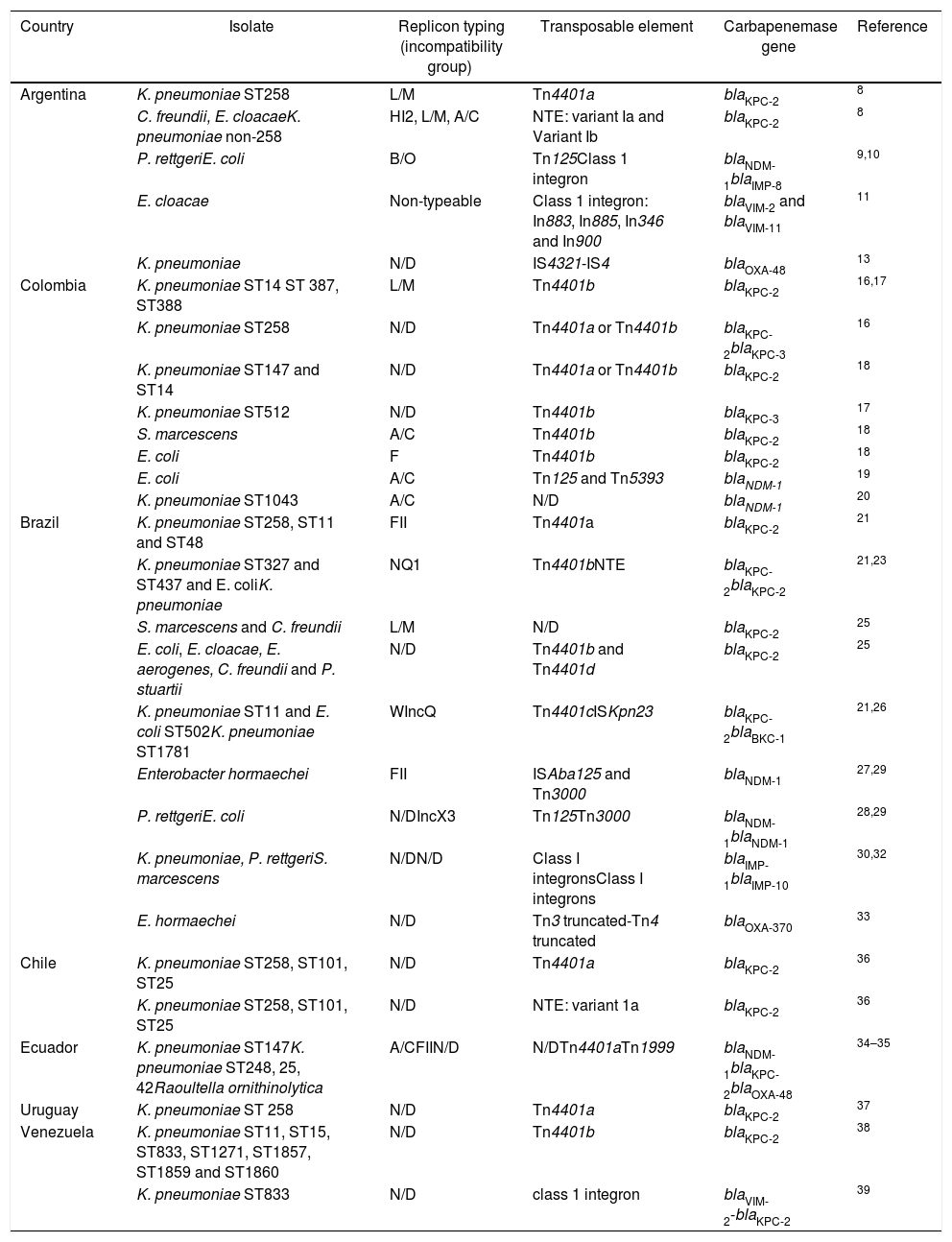
In South America, we observed the same patterns reported worldwide (Table 1), which indicates that some clones are being transmitted among South American countries (or elsewhere), raising the need for tracking carbapenemase clone dissemination. There is also evidence of carbapenemase gene mobility between plasmids and clones which potentially facilitates the emergence of new epidemic CRE clones such as K. pneumoniae ST11 and the hypervirulent clone ST25 (Table 1). This type of mobility is more difficult to track; plasmids carrying these genes could be identified (by replicon typing or plasmid/whole genome sequencing), but the rearrangements due to recombination and transposition sometimes complicate their traceability.8,42 A clear example of this evolution is the blaKPC-2 gene in K. pneumoniae CC258, which has been associated with IncFII derivatives such as pKpQIL43 and other plasmid incompatibility groups in Argentina, Brazil, Colombia, and Ecuador (Table 1) which have been recovered from K. pneumoniae CC258, non-CC258, and other members of the Enterobacterales.44,45 This variability of plasmids carrying similar blaKPC genetic environments suggests active plasmid rearrangements driven by transposons (e.g. Tn4401). The blaKPC-2 dissemination from the original host (K. pneumoniae) to other species of Enterobacterales is consequence of this evolutionary process: an active transposon harboring antimicrobial resistance genes finding broad (e.g. L/M, N, W) or wide host range plasmids (e.g. IncF, IncH), like it was found in South America (Table 1).46
Summary of mobile genetic elements reported in carbapenemase-producing Enterobacterales in South America.
| Country | Isolate | Replicon typing (incompatibility group) | Transposable element | Carbapenemase gene | Reference |
|---|---|---|---|---|---|
| Argentina | K. pneumoniae ST258 | L/M | Tn4401a | blaKPC-2 | 8 |
| C. freundii, E. cloacaeK. pneumoniae non-258 | HI2, L/M, A/C | NTE: variant Ia and Variant Ib | blaKPC-2 | 8 | |
| P. rettgeriE. coli | B/O | Tn125Class 1 integron | blaNDM-1blaIMP-8 | 9,10 | |
| E. cloacae | Non-typeable | Class 1 integron: In883, In885, In346 and In900 | blaVIM-2 and blaVIM-11 | 11 | |
| K. pneumoniae | N/D | IS4321-IS4 | blaOXA-48 | 13 | |
| Colombia | K. pneumoniae ST14 ST 387, ST388 | L/M | Tn4401b | blaKPC-2 | 16,17 |
| K. pneumoniae ST258 | N/D | Tn4401a or Tn4401b | blaKPC-2blaKPC-3 | 16 | |
| K. pneumoniae ST147 and ST14 | N/D | Tn4401a or Tn4401b | blaKPC-2 | 18 | |
| K. pneumoniae ST512 | N/D | Tn4401b | blaKPC-3 | 17 | |
| S. marcescens | A/C | Tn4401b | blaKPC-2 | 18 | |
| E. coli | F | Tn4401b | blaKPC-2 | 18 | |
| E. coli | A/C | Tn125 and Tn5393 | blaNDM-1 | 19 | |
| K. pneumoniae ST1043 | A/C | N/D | blaNDM-1 | 20 | |
| Brazil | K. pneumoniae ST258, ST11 and ST48 | FII | Tn4401a | blaKPC-2 | 21 |
| K. pneumoniae ST327 and ST437 and E. coliK. pneumoniae | NQ1 | Tn4401bNTE | blaKPC-2blaKPC-2 | 21,23 | |
| S. marcescens and C. freundii | L/M | N/D | blaKPC-2 | 25 | |
| E. coli, E. cloacae, E. aerogenes, C. freundii and P. stuartii | N/D | Tn4401b and Tn4401d | blaKPC-2 | 25 | |
| K. pneumoniae ST11 and E. coli ST502K. pneumoniae ST1781 | WIncQ | Tn4401cISKpn23 | blaKPC-2blaBKC-1 | 21,26 | |
| Enterobacter hormaechei | FII | ISAba125 and Tn3000 | blaNDM-1 | 27,29 | |
| P. rettgeriE. coli | N/DIncX3 | Tn125Tn3000 | blaNDM-1blaNDM-1 | 28,29 | |
| K. pneumoniae, P. rettgeriS. marcescens | N/DN/D | Class I integronsClass I integrons | blaIMP-1blaIMP-10 | 30,32 | |
| E. hormaechei | N/D | Tn3 truncated-Tn4 truncated | blaOXA-370 | 33 | |
| Chile | K. pneumoniae ST258, ST101, ST25 | N/D | Tn4401a | blaKPC-2 | 36 |
| K. pneumoniae ST258, ST101, ST25 | N/D | NTE: variant 1a | blaKPC-2 | 36 | |
| Ecuador | K. pneumoniae ST147K. pneumoniae ST248, 25, 42Raoultella ornithinolytica | A/CFIIN/D | N/DTn4401aTn1999 | blaNDM-1blaKPC-2blaOXA-48 | 34–35 |
| Uruguay | K. pneumoniae ST 258 | N/D | Tn4401a | blaKPC-2 | 37 |
| Venezuela | K. pneumoniae ST11, ST15, ST833, ST1271, ST1857, ST1859 and ST1860 | N/D | Tn4401b | blaKPC-2 | 38 |
| K. pneumoniae ST833 | N/D | class 1 integron | blaVIM-2-blaKPC-2 | 39 |
N/D: not determined.
Another approach to partially assess some of these complexities could be analyzing the genetic environment in which these genes are located. A classic example would be Tn4401, a Tn3-type, 10-kb mobile transposon frequently associated with blaKPC genes. Its structure consists of a Tn3 transposase gene (tnpA), a resolvase gene (tnpR) and additional insertion sequences (ISKpn6 and ISKpn7) delimited by two 39-bp imperfect inverted repeats (IRs).47 The isoforms (a to h) are classified based in nucleotide deletions upstream of blaKPC gene (affecting its promotor region and the KPC-expression), lacking genes or both: for instance, isoform “a” has a 99bp deletion; isoform “b” has no deletion; isoform “c”, a 215bp deletion; isoform “d”, a 68bp deletion; isoform “e”, a 255bp deletion; isoform “f”, a truncation in tnpA, absence of tnpR, ISKpn7 left part and Tn4401 IRL-1; isoform “g” is similar to “f” plus a 215bp deletion (like isoform c); and isoform “h” has a 188-bp deletion.48 (Fig. 1). Non-Tn4401 elements are other genetic environments of the blaKPC gene. These are complex DNA structures sharing some Tn4401 elements such as ISKpn6. Some examples are shown in Fig. 1.11,49,50 In South America the isoforms Tn4401a and b were the most commonly found in regional studies (e.g. in Argentina, Colombia, Brazil, Chile, Ecuador, and Venezuela), but NTEs and the isoforms Tn4401c and d were also described in Brazil, respectively, mainly in non-K. pneumoniae isolates (Table 1). As mentioned before, this lateral dissemination between genera and species would be more related to the type of conjugative plasmid where the transposon ‘landed’.
Currently, more than 70 MBLs (chromosomally- and plasmid-encoded) have been reported and grouped based on DNA sequence similarity.51 NDM is one of the most successfully plasmid-disseminated MBL detected in different members of the Enterobacterales around the world and their frequent association with ISAba125 suggests a possible Acinetobacter spp. origin, a bacterium in which this association is very common.52 Tn125, an ISAba125-based composite transposon, was one of the genetic elements described to be involved in blaNDM dissemination, but in Enterobacteriaceae Tn125 it has been interrupted or truncated, generating a variety of different genetic contexts for blaNDM.53K. pneumoniae, E. coli, and Enterobacter spp. are the predominant carriers of blaNDM, with heterogeneous clonal backgrounds and multiple acquisitions of blaNDM genes across bacterial species, also, different replicon types of blaNDM-carrying plasmids in the Enterobacteriaceae were described, being the most common IncA/C, FIA, FIB, FII and X353 (in our bibliographic review we found less frequent descriptions of MBL-producing Enterobacteriaceae). Reported blaNDM-1 genes were associated with ISAba125 composite transposon Tn125 (Table 1). There are no extensive descriptions of bacterial clones and plasmid incompatibility groups related to blaNDM in South America. In Argentina, there was one description of a blaNDM-1-harboring, IncB/O plasmid recovered from P. rettgeri; in Colombia and Ecuador, blaNDM-1 was detected on IncA/C plasmids from E. coli (Colombia) and K. pneumoniae (both countries). More information has to be developed and analyzed to have a clearer picture of blaNDM spread in South America. A similar scenario was found regarding blaVIM (Argentina-Venezuela) and blaIMP (Brazil), both carbapenemase genes associated to class 1 integrons, also described in Pseudomonas aeruginosa, evidencing that class I integrons are efficient genetic platforms to incorporate MBLs genes and disseminated once a transposon or plasmid is involved.54,55
The OXA-enzymes have higher hydrolysis rates against cloxacillin and oxacillin than other β-lactams. In addition, they are poorly inhibited by clavulanic acid, tazobactam, and aztreonam, and some can inactivate carbapenems.56 We found few descriptions of MGEs associated with these carbapenemase genes in South America; in Ecuador, the blaOXA-48-like gene was related to Tn1999-like.57 However, in Brazil blaOXA-347, a variant of blaOXA-48, showed association with truncated Tn3 and Tn4 transposons.
ConclusionsCurrent evidence enhances the importance of MGEs for carbapenemase gene dissemination. Except for Argentina, Brazil, and Colombia, the reports of MGEs are scarce in other South American countries. The blaKPC and blaNDM are the most prevalent carbapenemase genes reported in Enterobacterales species are associated with Tn4401/non-Tn4401 elements and ISAba125/Tn125 respectively while blaVIM and blaIMP carbapenemase genes are related to class 1 integrons. The location of transposons or integrons in different plasmid incompatibility groups and bacterial clones denotes their capacity in transferability and mobilization. It is possible that intercontinental dissemination of CRE clones was followed by horizontal gene transfer of plasmids carrying carbapenem resistance genes to local bacteria. Any control measure intended to tackle antibiotic resistance genes dissemination requires the identification of the potential sources of these genes, which nowadays is carried out by DNA polymorphisms. Unfortunately, carbapenem resistance genes have very little polymorphisms. We argue that the study of MGEs associated with these genes could provide very valuable epidemiological information to detect the potential sources.
Funding informationThis study was funded by Instituto de microbiología, Universidad San Francisco de Quito.
Conflict of interestThe authors declare no conflicts of interest.