The Mycobacterium tuberculosis East African-Indian (EAI) spoligotyping family (belonging to lineage 1, Indo-Oceanic, defined by the region of deletion RD239) is distributed worldwide, but is more prevalent in Southeast Asia, India, and East Africa. Studies in Latin America have rarely identified EAI. In this study, we describe the occurrence of the EAI family in Brazil.
MethodsEAI was identified in a systematic literature review of genetic diversity studies pertaining to M. tuberculosis in Brazil, as well as in a survey conducted in Salvador, Bahia, located in the northeastern region of this country.
ResultsThe EAI6-BGD1 spoligotyping family and the EAI5 Spoligotype International Type (SIT) 1983 clade were the most frequently reported, with wide distribution of this particular clade described in Brazil. The distribution of other EAI spoligotyping patterns with broader worldwide distribution was restricted to the southeastern region of the country.
ConclusionsEAI may be endemic at a low frequency in Brazil, with some clades indicating increased fitness with respect to this population.
Tuberculosis (TB) remains one of the world deadliest communicable diseases.1Mycobacterium tuberculosis (Mtb) is broadly distributed across all five continents, although relatively few countries account for as much as 80% of all TB cases.1 To date, seven lineages of Mtb complex have been recognized and associated with particular geographical regions.2 There is evidence suggesting that these phylogeographic groups differ in their biological fitness and are best adapted to the sympatric human hosts.3,4
Three major spoligotyping families are most frequently found in Africa, Central America, Europe, and South America: Haarlem, Latin American-Mediterranean (LAM) and T from lineage 4 (which is defined by two deletions in the genome, one comprising the TbD1 region and another at the pks15/1 locus).2,5–7 The East African-Indian (EAI) spoligotyping family from lineage 1 (defined by the region of deletion RD239)2,5–7 is prevalent in Southeast Asia, particularly in the Philippines, Myanmar, Malaysia, Vietnam, Thailand, India, and East Africa, yet is relatively rare in the Americas.7
EAI, a mildly virulent group, possesses a reduced potential for transmissibility.8 Studies performed to characterize Mtb strains in circulation in Latin America have rarely identified EAI, and this family is considered to have limited distribution in comparison with other families in this region.7 Here we report the detection of one case of the EAI family in Salvador, Bahia-Brazil, an area endemic for TB, and we review previous reports of the occurrence of this family in Brazil.
MethodsRecruitment and study designA population-based study conducted in Salvador, the capital of the state of Bahia, from August 2008 to August 2010 involved sputum smear-positive patients in the context of an epidemiological survey that identified patients with pulmonary tuberculosis at local health clinics, with enrollment limited to one person per household (data not published). This is an endemic area where disease incidence was estimated at 62.7/100,000 in 2014, while the overall TB incidence in Brazil was reported to be 33.5/100,000.9 A total of 362 mycobacterial isolates were obtained from the collected positive sputum smears, among which 351 isolates were successfully genotyped and one isolate was identified as belonging to the EAI family. We describe this finding herein and review the previous occurrence of EAI in the country. The present study (CAAE: 0016.0.069.000-07) was approved by the Ethics Committee of the Gonçalo Moniz Research Center (Fiocruz). It adhered to Resolution 196/96 established by the Brazilian National Health Council and complied with the Helsinki declaration guidelines.
Species identification and genotypingIdentification at the species level was performed by phenotypic and biochemical methods after culturing in Lowenstein-Jensen medium (Becton-Dickinson, Palo Alto, CA). RFLP was performed according with the method described by van Embden et al.10 Spoligotyping profiles were obtained using the method established by Cowan et al.11 and then submitted to the SITVIT WEB database for family and subfamily designation.7 Single Nucleotide Polymorphisms (SNPs) were genotyped according to the method by Lopes et al.12 using 59 SNPs located outside the genome regions known to be related to antibiotic resistance. The EAI lineage has been previously defined by the SNPs in Rv1020_256 and Rv2362c_606.13
Systematic review of the literatureWe reviewed the studies of Mtb population genetics performed in Brazil to identify previous findings of EAI family tuberculosis isolates in the country. Published studies were located using the PubMed platform or the electronic libraries Scielo (Scientific Electronic Library Online Brazil) or BIREME (Virtual Health Library) through searches using the following terms: “M. tuberculosis” and (genotyping or spoligotyping) and Brazil, imposing no language restrictions. Studies were included in the analysis if fulfilling the following criteria: (i) Study reports more than 10 tuberculosis cases from Brazil; (ii) Study discriminates lineages of Mtb and identifies EAI family by spoligotyping or SNP; (iii) Population-based study published up to June 2016.
Designation of phylogenetic groups and genetic similarity analysesThe octal or binary spoligotyping patterns reported were retrieved from the articles obtained with this search and submitted (1) to the SITVIT WEB database for family and subfamily designation; and (2) to the MIRU-VNTRplus database to generate neighbor-joining phylogenetic trees either including or not the reference strains from this web application. If the SIT information was not available and neither the octal nor the binary spoligotyping pattern was described in the article, the strain was not used in this phylogenetic analysis.
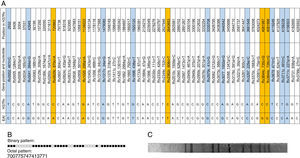
ResultsIn the survey conducted in Salvador, Bahia, only one case of EAI (0.3%) was identified among the 351 successfully genotyped isolates. This corresponded to a 31-year-old diabetic male presenting characteristic symptoms of TB: cough, hemoptysis, night sweats and weight loss. This patient was interviewed and reported that he had never changed residence, nor traveled outside the metropolitan area. He also reported that, in his adolescence, he had contact with a visiting relative from Italy who exhibited typical TB symptoms. The isolate retrieved from this patient was assigned to the EAI6-BGD1 SIT 702 clade in accordance with the spoligotyping profile observed (Fig. 1B). This unique EAI strain was identified by 15 of the 59 SNPs investigated in our series (Table 1, Fig. 1A). Lopes et al.12 showed that although 10 of these polymorphisms occur in other lineages, the presence of five SNPs: Rv 0629c_0870, Rv 1020_0256, Rv 2362c_0606, Rv 3644c_0726, and Rv 3644c_0735, serves as confirmation of EAI. The RFLP pattern showed 12 bands (Fig. 1C) and was unique in our series (data not published).
Genotypic profile of the isolate from Salvador, Bahia, Brazil assigned to the EAI6-BDG1 spoligotyping subfamily (SIT 702). (A) Single Nucleotide Polymorphisms (SNPs). Loci highlighted in blue show polymorphisms uniquely identified in the East African-Indian (EAI) strain as compared to the other 351 genotyped isolates in this series. Loci highlighted in orange show the SNPs that were also distinctive of EAI, according to Lopes et al.12 (B) Spoligotyping pattern. (C) Restriction-fragment length polymorphism (RFLP).
Panel of 59 single nucleotide polymorphisms (SNPs) used for genotyping.
| Genome location | Gene | Gene position and nucleotide | S or NS | Reference |
|---|---|---|---|---|
| 2532 | Rv0002 | Rv0002_481t>C | S | 13 |
| 6406 | Rv0005 | Rv0005_1284c>T | S | 32 |
| 9304 | Rv0006 | Rv0006_2003g>A | NS | 33 |
| 37031 | Rv0034 | Rv0034_165c>G | S | 34 |
| 43945 | Rv0041 | Rv0041_384a>G | S | 34 |
| 92199 | Rv0083 | Rv0083_188t>G | S | 34 |
| 157292 | Rv0129c | Rv0129c_309g>A | S | 33 |
| 220050 | Rv0189c | Rv0189c_1674g>A | S | 34 |
| 311613 | Rv0260c | Rv0260c_1047c>A | S | 34 |
| 720863 | Rv0629c | Rv0629c_870c>A | S | 13 |
| 797736 | Rv0697 | Rv0697_804c>T | S | 34 |
| 918316 | Rv0824c | Rv0824c_435a>G | S | 34 |
| 923065 | Rv0831c | Rv0831c_645a>T | S | 34 |
| 1047683 | Rv0938 | Rv0938_1548g>T | NS | 35 |
| 1068151 | Rv0956 | Rv0956_591t>C | S | 34 |
| 1139222 | Rv1020 | Rv1020_256g>A | NS | 13 |
| 1163134 | Rv1040c | Rv1040c_243a>G | S | 34 |
| 1178116 | Rv1056 | Rv1056_489t>C | S | 34 |
| 1477588 | Rv1316c | Rv1316c_44c>G | NS | 13 |
| 1479085 | Rv1317c | Rv1317c_34a>G | NS | 13 |
| 1548149 | Rv1375 | Rv1375_318G | S | 34 |
| 1588456 | Rv1411c | Rv1411c_27t>C | S | 33 |
| 1595342 | Rv1420 | Rv1420_1301t>C | NS | 13 |
| 1884697 | Rv1662 | Rv1662_2994G>a | S | 34 |
| 1892017 | Rv1665 | Rv1665_792t>C | S | 34 |
| 1920120 | Rv1696 | Rv1696_438g>T | NS | 13 |
| 1960391 | Rv1733c | Rv1733c_97c>T | NS | 33 |
| 2134215 | Rv1884c | Rv1884c_47a>G | S | 33 |
| 2239349 | Rv1996 | Rv1996_346a>G | NS | 33 |
| 2278276 | Rv2030c | Rv2030c_111c>T | S | 33 |
| 2603797 | Rv2330c | Rv2330c_426c>T | S | 33 |
| 2627946 | Rv2349c | Rv2349c_753T>c | S | 34 |
| 2643653 | Rv2362c | Rv2362c_606c>T | S | 13 |
| 2825581 | Rv2510c | Rv2510c_1509a>C | S | 36 |
| 2880702 | Rv2560 | Rv2560_628g>C | NS | 34 |
| 2891267 | Rv2567 | Rv2567_1473c>T | S | 34 |
| 3300104 | Rv2949c | Rv2949c_467g>A | NS | 33 |
| 3300196 | Rv2949c | Rv2949c_375c>T | S | 33 |
| 3312632 | Rv2959c | Rv2959c_207g>A | NS | 33 |
| 3332254 | Rv2976c | Rv2976c_501g>A | S | 13 |
| 3335708 | Rv2979c | Rv2979c_41c>G | NS | 13 |
| 3426795 | Rv3062 | Rv3062_1212c>G | S | 13 |
| 3438386 | Rv3075c | Rv3075c_588c>T | S | 34 |
| 3440542 | Rv3077 | Rv3077_1002a>G | S | 34 |
| 3455686 | Rv3088 | Rv3088_1347g>C | S | 34 |
| 3544710 | Rv3176c | Rv3176c_591a>G | S | 34 |
| 3597737 | Rv3221c | Rv3221c_30g>A | S | 33 |
| 3641447 | Rv3261 | Rv3261_905c>T | NS | 33 |
| 3681548 | Rv3297 | Rv3297_229a>C | S | 13 |
| 3783058 | Rv3370c | Rv3370c_1683c>T | S | 34 |
| 4024273 | Rv3581c | Rv3581c_75a>G | S | 34 |
| 4081987 | Rv3644c | Rv3644c_735c>G | S | 13 |
| 4081996 | Rv3644c | Rv3644c_726c>G | S | 13 |
| 4119246 | Rv3679 | Rv3679_471T>c | S | 34 |
| 4137829 | Rv3695 | Rv3695_624c>T | S | 34 |
| 4156239 | Rv3711c | Rv3711c_491t>C | NS | 13 |
| 4156503 | Rv3711c | Rv3711c_227g>A | NS | 13 |
| 4182695 | Rv3731 | Rv3731_938g>A | NS | 13 |
| 4255922 | Rv3799c | Rv3799c_27t>C | S | 34 |
S, synonymous; NS, non-synonymous.
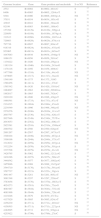
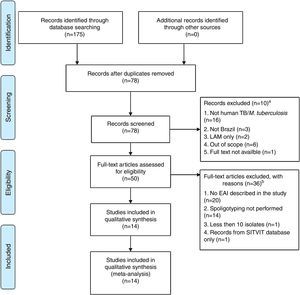
Our systematic review of the literature regarding EAI occurrence in studies of Mtb diversity performed in Brazil yielded 175 articles, of which 14 were considered eligible for analysis (Fig. 2 and Table 2).14–27 Most of these were either based on bacterial collections maintained in reference laboratories that perform culturing for species identification and phenotypic drug-susceptibility testing,14,16–18,20,21,24,26,27 or on convenience sampling of TB patients at reference health care units responsible for TB diagnosis.15,22,23,25 One report consisted of a case-control study involving drug-sensitive versus drug-resistant TB patients.19
PRISMA31 flow diagram describing the systematic literature review performed. aRecords screened were excluded after reading the title and abstract if: (i) the study did not focus on M. tuberculosis; (ii) the isolates were not identified in Brazil; (iii) the strains analyzed were restricted to a non-East African-Indian (EAI) family of M. tuberculosis; (iv) the study did not focus on isolates from humans; (v) the full text was not available via the CAPES Consortium, or access to the article was not provided by Fiocruz. bFull-text articles were not included if: (i) they did not report EAI; (ii) spoligotyping was not performed; (iii) less than 10 isolates were described; (iv) the study analyzed records exclusively from the SITVIT database.
Literature review summary of East African-Indian (EAI) isolates described in studies of Mycobacterium tuberculosis diversity performed in Brazil.
NA, not available.
aAmbiguous.
bPattern retrieved from the SITVIT WEB based on the SIT informed by the authors.
cAs described by the authors.
dNo match was found in the SITVIT WEB database; the closest matches found in the MIRU-VNTRplus database pertain to the LAM family.
The EAI family is rare in Brazil, occurring typically at frequencies below 2% of the datasets analyzed (Table 2). While EAI family isolates were found in the North,22,24 Northeast,19,22 South,14,16,26 and Southeast of Brazil,15,17,18,20–23,27 the highest circulation of this family was reported in the North (Pará)22 (Table 2). The number of EAI isolates varied from only one to as many as 15 in the studies reviewed (Table 2).
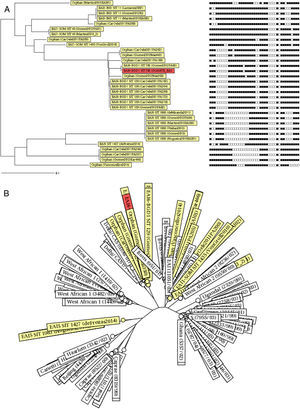
The EAI subfamilies most frequently reported in Brazil were EAI6-BGD1 (especially in Pará,22,24 SIT 129) and EAI5 (SIT 1983) (Fig. 3). Some orphan patterns similar to EAI6-BGD1 were also described in two studies performed in Pará (Fig. 3A). Strains of the EAI5 subfamily SIT 1983 were consistently reported (Table 2 and Fig. 3A).14,20–23 Furthermore, the spoligotyping patterns EAI3-IND SIT 11 and EAI1-SOM SIT 48 were also present in more than one study (Table 2 and Fig. 3A). These spoligotyping patterns are more similar to other EAI reported in the MIRU-VNTRplus database,28,29 while the EAI6-BGD1 isolates described in Pará and Bahia cluster with the Delhi/Central Asian (CAS) spoligotype (Fig. 3B).
Neighbor-joining trees depicting similarities between the East African-Indian (EAI) spoligotyping patterns in Brazil retrieved from the systematic review of the literature and the spoligotyping pattern described in Salvador, Bahia. (A) Dendrogram with corresponding binary spoligotyping patterns. (B) Radiation tree including the reference strains from the MIRU-VNTRplus database.
The low frequency of EAI in Brazil suggests lower transmissibility of this phylogeographic group than what is observed in other Mtb families. Moreover, other authors have argued that immigration has resulted in the steady influx of particular EAI strains, which has been identified throughout Brazil. On the other hand, this broad distribution, taken together with the restricted genetic variability of the EAI isolates identified in the country, may indicate the endemic nature of this family, albeit at a low prevalence.
The most prevalent subfamilies found in Brazil were EAI6-BGD1 and EAI5. Despite the fact that these subfamilies include spoligotype patterns that are common worldwide, the EAI6-BGD1 SIT 702 and the EAI5 SIT 1983 clades described herein have restricted circulation outside Brazil. The SITVIT WEB database contains 21 isolates with the EAI6-BGD1 SIT 702 pattern, three of which are Brazilian samples from an outbreak in Pará.7,22 The remaining specimens were isolated in Cuba, French Guiana (from a patient of Brazilian origin), the United Kingdom, Malawi, Tunisia, and Zambia.7 For SIT 1983, the clade with the widest distribution in Brazil, the SITVIT WEB database contains data only from Brazil and India.7 Moreover, strains of the EAI6-BGD1 family (as well as orphan spoligotyping patterns similar to this subfamily, so far not described in the SITVIT WEB database) were previously reported in an outbreak in Pará,22,24 as well as in our series in Salvador, Bahia, which is indicative of ongoing transmission not restricted to a particular setting. While the EAI6-BGD1 SIT 129 pattern described in Pará has broader worldwide distribution, as isolates from this clade have been previously reported outside Brazil in Germany, the Republic of Congo, Malawi, Zimbabwe, South Africa, Zambia, French Guiana, and the United States,7 it was also identified in the context of the Pará outbreak. Finally, other widely distributed EAI clades, such as EAI3-IND SIT 11 and EAI1-SOM SIT 487 (as well as highly similar orphan patterns) were exclusively found in studies performed in southeastern Brazil. Taken together, these findings suggest that some specific clades of EAI may be better adapted to particular Brazilian populations.
Interestingly, EAI6-BGD1 and similar orphan spoligotyping patterns described in Salvador-Bahia and in the state of Pará, as well as the isolates obtained from the outbreak that occurred in this state, cluster with some strains of lineage 3 (defined by the combined deletion of the genomic regions TbD1 and RD750, including the Delhi/Central Asian (CAS) spoligotyping family2,5–7). Lineage 3 belongs to a group of modern Mtb lineages considered to be more virulent than EAI.2,5–7 Nonetheless, this finding should be interpreted with caution, due to the limited capacity of spoligotyping to accurately distinguish among monophyletic groupings of Mtb.30
ConclusionsIn spite of the low detected prevalence, EAI may in fact be endemic in Brazil. The restricted worldwide distribution of some spoligotyping patterns described in multiple studies conducted in Brazil, together with the genetic relatedness found among isolates from different parts of the country and the occurrence of an outbreak in Pará, seem to suggest the increased fitness exhibited by some clades with respect to our population.
FundingConselho Nacional de Desenvolvimento Científico e Tecnológico (Edital MCT-CNPq/MS-SCTIE-DECIT – N° 25/2006).
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank João Costa and Isabel Marques for their excellent technical assistance, as well as Andris Walter for expert language review of the manuscript.