In the pandemic, rapid and accurate detection of SARS-CoV-2 is crucial in controlling the outbreak. Recent studies have shown a high detection rate using saliva/oral fluids as specimens for laboratory detection of the virus. We intended to evaluate the test performance of the Xpert Xpress SARS-CoV-2 cartridge assay in comparison to a conventional qRT-PCR testing, using saliva as biological specimen. Forty saliva samples from symptomatic participants were collected. Conventional qRT-PCR was performed for amplification of E and RdRp genes and the Xpert Xpress SARS-CoV-2 assay amplified E and N2 genes. In the conventional assay, the median cycle threshold value of the E gene was 34.9, and of the RdRp gene was 38.3. In the Xpert Xpress assay, the median cycle threshold value of the E gene was 29.7, and of the N2 gene was 31.6. These results can allow a broaden use of molecular tests for management of COVID-19 pandemic, especially in resources-limited settings.
Since the beginning of SARS-CoV-2 pandemic, the number of infected people is getting higher. According to World Health Organization (WHO), by October 22th, 2020, a total of 40,890,712 confirmed cases of COVID-19, including 1,126,351 deaths was reported.1 Rapid and accurate detection of SARS-CoV-2 is an essential step in controlling the outbreak, and multiple diagnostic tests, including antigen detection, serological and molecular assays, have been rapidly developed.2
Real Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR) is the gold standard test for detection of SARS-CoV-2, whether using in house, fully automated or cartridge-based assays. However, such molecular assays use respiratory specimens (oropharyngeal and nasopharyngeal swabs, bronchoalveolar wash, tracheal aspirate).2 Collecting nasopharyngeal swab (NPS)/oropharyngeal swab (OPS) causes discomfort to patients due to invasiveness of the procedure, which can reduce the possibility of patient consent to retest, and may represent a considerable risk for healthcare workers, because of its potential to induce patients to sneeze or cough, generating virus-containing aerosols.3
Recent studies have shown a high detection rate using saliva/oral fluids as specimens for laboratory detection of SARS-CoV-2.3–6 The use of saliva as biological sample has several advantages, such as easy self-collection even at home, and no need of trained personnel for sample collection. In addition, saliva collection is much more comfortable for the patient than NPS or OPS procedures. It also saves time, and is less costly, because it does not require the use of personal protective equipment nor viral transportation solution.5
The aim of this study is to evaluate the test performance of the Xpert Xpress SARS-CoV-2 cartridge assay compared to a qRT-PCR testing, using saliva as biological specimen.
This study was developed at the Infectious Diseases Research Laboratory (LAPI), of Complexo Hospitalar Professor Edgard Santos (C-HUPES), Federal University of Bahia, in Salvador, Brazil. We used 40 saliva samples collected during June and July 2020, from a previous validation study from our group.6 Saliva samples from symptomatic participants were collected into 30 mL sterile urine cups, and kept at −80 °C until testing. Participants were instructed to repeatedly spit until approximately 2 mL of sample was obtained, thus avoiding mucous secretions from oropharynx or lower respiratory tract (i.e., sputum). Samples were diluted 1:1 with phosphate buffered saline (PBS) 1×. This study was approved by the Research Ethics Committee of Maternidade Climério de Oliveira–UFBA (approval number: 4.042.620). Informed consent was obtained from all participants enrolled in this study.
RNA isolation was performed by using viral RNA mini kit (QIAGEN, Hilden, Germany), according to manufacturer’s instructions. RNA template was subjected to conventional qRT-PCR amplification according to Charité-Berlin protocol,7 which consists of detection of envelope gene E and Orf1ab RdRp gene. Amplification reactions were carried out on Applied Biosystems 7500 Real Time PCR detector, and results were classified as positive for SARS-CoV-2 when both E and RdRp genes were detected and cycle threshold (Ct, number of cycles required for the fluorescent signal to exceed background level) values were less than 40.9.
The Xpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, California, USA) is a very recently released assay for use under the Emergency Use Authorization (EUA) only, from U.S. Food and Drug Administration (FDA). The assay is a sample-to-answer qRT-PCR test, requires a sample load of 300 µL, has a detection limit of 250 copies per mL, and a running time of approximately 45 min. The target genes are E and nucleocapsid N2 genes. The detection of both targets or N2 alone is considered positive, and the detection of E gene alone is considered presumptive positive (Ct value<45).8
Statistical analyses were performed using Statistical Package for the Social Sciences software (SPSS) version 18.0. Linear regression analysis was used to calculate R2.
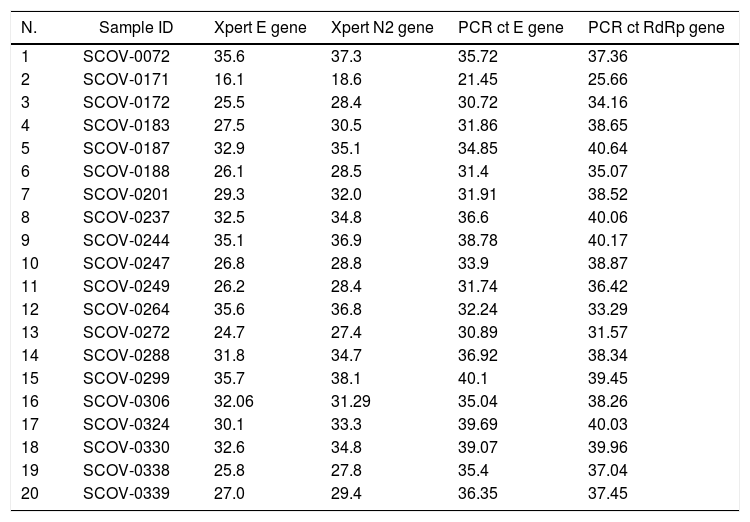
We selected, 20 positive and 20 negative saliva samples already tested by the conventional qRT-PCR assay (Charité–Berlin test) to be validated in the Xpert Xpress SARS-CoV-2 system. A detailed comparison of the assay results is shown in Table 1.
Positive samples Ct values for the two different molecular assays, by the target gene of SARS-CoV-2.
| N. | Sample ID | Xpert E gene | Xpert N2 gene | PCR ct E gene | PCR ct RdRp gene |
|---|---|---|---|---|---|
| 1 | SCOV-0072 | 35.6 | 37.3 | 35.72 | 37.36 |
| 2 | SCOV-0171 | 16.1 | 18.6 | 21.45 | 25.66 |
| 3 | SCOV-0172 | 25.5 | 28.4 | 30.72 | 34.16 |
| 4 | SCOV-0183 | 27.5 | 30.5 | 31.86 | 38.65 |
| 5 | SCOV-0187 | 32.9 | 35.1 | 34.85 | 40.64 |
| 6 | SCOV-0188 | 26.1 | 28.5 | 31.4 | 35.07 |
| 7 | SCOV-0201 | 29.3 | 32.0 | 31.91 | 38.52 |
| 8 | SCOV-0237 | 32.5 | 34.8 | 36.6 | 40.06 |
| 9 | SCOV-0244 | 35.1 | 36.9 | 38.78 | 40.17 |
| 10 | SCOV-0247 | 26.8 | 28.8 | 33.9 | 38.87 |
| 11 | SCOV-0249 | 26.2 | 28.4 | 31.74 | 36.42 |
| 12 | SCOV-0264 | 35.6 | 36.8 | 32.24 | 33.29 |
| 13 | SCOV-0272 | 24.7 | 27.4 | 30.89 | 31.57 |
| 14 | SCOV-0288 | 31.8 | 34.7 | 36.92 | 38.34 |
| 15 | SCOV-0299 | 35.7 | 38.1 | 40.1 | 39.45 |
| 16 | SCOV-0306 | 32.06 | 31.29 | 35.04 | 38.26 |
| 17 | SCOV-0324 | 30.1 | 33.3 | 39.69 | 40.03 |
| 18 | SCOV-0330 | 32.6 | 34.8 | 39.07 | 39.96 |
| 19 | SCOV-0338 | 25.8 | 27.8 | 35.4 | 37.04 |
| 20 | SCOV-0339 | 27.0 | 29.4 | 36.35 | 37.45 |
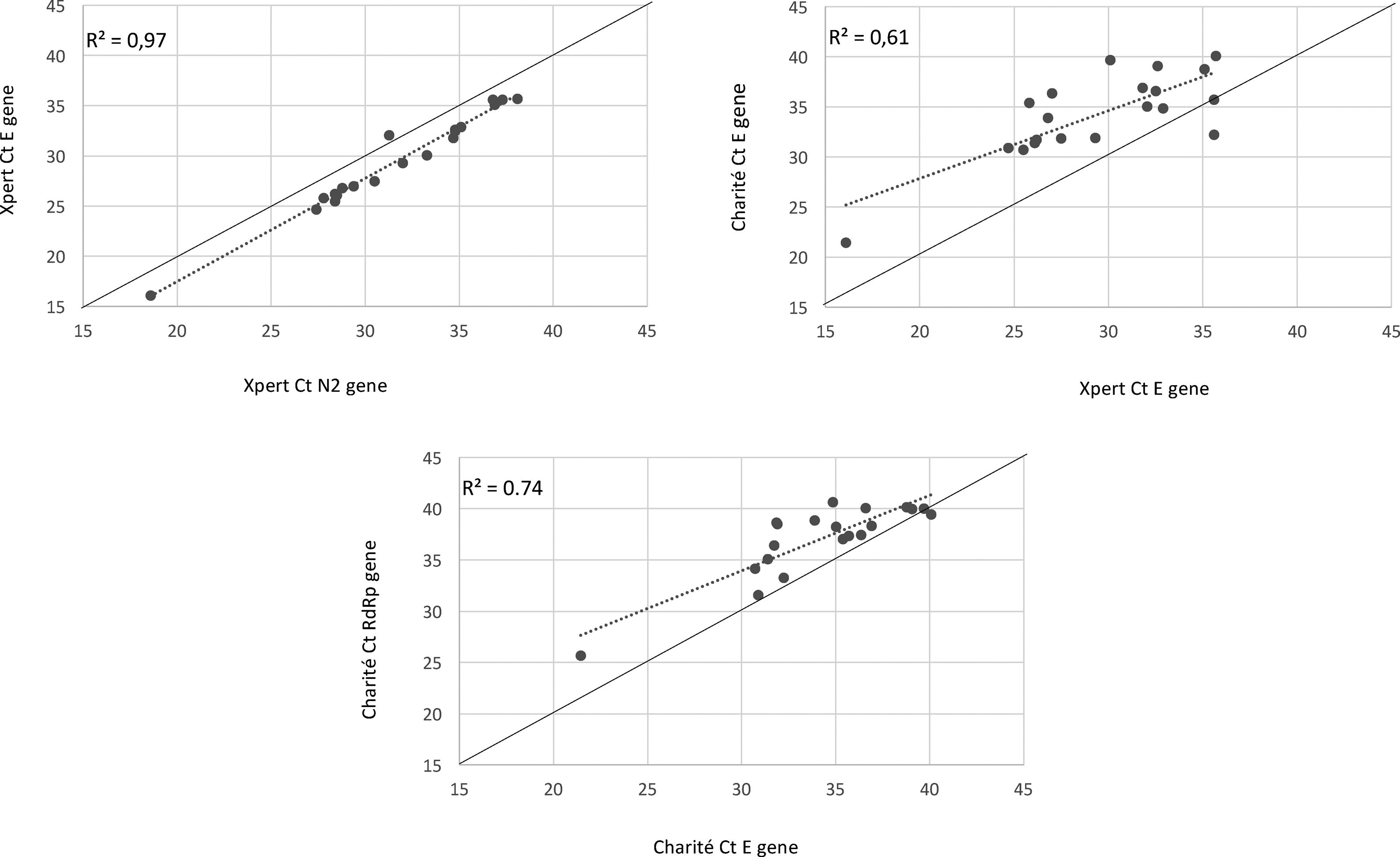
We observed 100% concordance between both assays. There were no presumptive positive results in the Xpert Xpress assay. In the Charité-Berlin protocol the median (IQR) Ct values of the E gene was 34.9 (31.8–36.8), and of the RdRp gene was 38.3 (35.4–39.8), while in the Xpert Xpress assay the median (IQR) Ct values of the E gene was 29.7 (26.1–32.8), and of the N2 gene was 31.6 (28.4–35.0). Fig. 1 shows the correlations (R2 values) between Ct values for both assays and for the different viral gene targets.
In a previous study we validated the use of saliva as a biological sample for the diagnosis of COVID-19. Results of conventional NPS and/or OPS versus saliva samples testing were compared by performing qRT-Real Time PCR assay.6 There was an overall high agreement (96.1%) between the two tests. In the present study, we detected an overall equivalence between results of qRT-Real Time PCR and Xpert Xpress SARS-CoV-2 assay.
The use of the Xpert Xpress SARS-CoV-2 assay was previously validated and results showed a great concordance over a range of SARS-CoV-2 viral loads and across established human coronaviruses.9,10 However, the samples used for assay validation were the conventional NPS/OPS. On the other hand, McCormick-Baw et al.11 have tested a total of 156 paired NPS and saliva specimens and found 98% of agreement.
Our results demonstrated that even samples presenting high Ct values (low viral load) in the conventional qRT-Real Time assay were detected in this cartridge-based assay. Similar results were presented by Broder et al.,12 but also using NPS/OPS.
The detected higher R2 value in Xpert E and N2 genes (0.97) compared to Charité RdRp and E genes (0.74) can demonstrate a lower test variability of an automated nucleic acid amplification test than conventional RT-PCR, as shown in a recently published study from Loeffelholz et al.13
Our work presents some limitations including the impossibility of testing viscous samples. Some saliva samples presenting excess of mucus were not included in this validation study. Only the liquid non-viscous components of each specimen were drawn into the disposable pipets for test cartridge inoculation.8 Although viscous saliva specimens could be treated with Sputasol (Thermo Scientific) or acetylcysteine to liquify the sample, we did not test this strategy in our study.
In conclusion, the Xpert Xpress system allows extensive testing for SARS-CoV-2 outside of the clinical laboratory environment, due to its easy handling. In addition, the use of self-collected saliva specimen is an easy, convenient, and low-cost alternative to conventional NP swab-based molecular tests. These results can allow a broaden use of molecular tests for management of COVID19 pandemic, especially in resources-limited settings.
Conflicts of interestThe authors declare no conflicts of interest.