The diagnosis of extrapulmonary tuberculosis is still a challenge because of its pauci-bacillary nature. The aim of the study was to evaluate the role of a multiplex PCR assay in the diagnosis of extrapulmonary tuberculosis and to compare the efficiency of two targets, IS6110 and MPB64 to detect Mycobacterium tuberculosis.
Methods150 extrapulmonary samples (61 pus/aspirate, 46 tissue, 32 body fluids, and 11 urine) from clinically suspected cases of tuberculosis were included in the study. All the samples were subjected to direct fluorescent microscopy, TB culture (BacT/ALERT 3D, biomerieux, Durham, North Carolina, USA) and a Multiplexed Tandem PCR targeting two mycobacterial DNA sequences, IS6110 and MPB64. Master-Mix reagents and primers were prepared by AusDiagnostics Pvt. Ltd (Alexandria, New South Wales, Australia). The performance of the assay was assessed using a composite gold standard, which included clinical characteristics, microbiology smear as well as culture, histopathology, cytology, radiology, and response to antitubercular therapy.
Results20.3%, 23.6%, and 45.3% of specimens were positive by smear, culture, and PCR, respectively. The sensitivity and specificity of the multiplex PCR was 91.9% and 88.4%, respectively, using the composite gold standard. Positive and negative predictive values of the PCR were estimated as 85.1% and 93.8%, respectively. Higher positivity was observed with target IS6110 (44.6%) as compared to target MPB64 (18.9%). The sensitivities of IS6110 and MPB64 individual targets were 90.3% and 64.5%, respectively, and specificities were 88.4% and 97.7%, respectively.
ConclusionPCR can play an important role in rapid and accurate diagnosis of extrapulmonary tuberculosis. IS6110 alone is an effective target in our part of the country.
India is a high tuberculosis (TB) burden country contributing to 26% of the global burden of the disease. While pulmonary TB (PTB) is the most common form of disease, extrapulmonary tuberculosis (EPTB) constitutes around 20% of all TB cases in India.1 Early diagnosis is essential in instituting effective and timely therapy. Conventional methods like smear and culture are of limited use in diagnosis of EPTB cases due to low sensitivity. Amplification techniques have attracted considerable interest in the diagnosis of tuberculosis particularly with the hope of shortening the time required in detection. Most of the assays are evaluated for pulmonary specimens and smear positive specimens. The diagnosis of extrapulmonary and smear negative tuberculosis is still a challenge especially because of the pauci-bacillary nature. Despite numerous reports in literature, amplification techniques do not yet have an established role in the laboratory diagnosis of tuberculosis, nor have they replaced traditional techniques. Some studies have evaluated the role of PCR in the diagnosis of EPTB using various primers to amplify different targets.2–4 Highly conserved insertion sequences, IS6110, is the most commonly used target in the detection of Mycobacterium tuberculosis. Few Indian studies have pointed out that the most commonly used method targeting insertion sequence IS6110 has a danger of missing at least 10% of Indian tuberculosis patients harboring zero copy number of IS6110.5 Therefore, there is a need to know which other targets can be used to reliably detect the EPTB cases. The other candidate target used by the researchers in India has been the gene encoding mpt64 (MPB64), a secretary mycobacterial antigen present only in M. tuberculosis complex, for the diagnosis of TB.6,7 In the present study the comparative value of a multiplex PCR system which simultaneously amplifies two targets, for their role in the early detection of EPTB has been analyzed using a multiplex real-time PCR targeting IS6110 and MPB64. Keeping in view the poor rate of culture positivity in EPTB which makes it difficult to evaluate assays when it is considered as the gold standard, we have also made an attempt to evaluate this multiplex PCR using a composite gold standard (CGS) based on clinical presentation along with radiology, histopathology, and response to anti-tuberculous therapy. Further, the present study undertook to evaluate the role of a multiplex real-time PCR assay in the laboratory diagnosis of EPTB in comparison to conventional bacteriological techniques like smear and culture.
Material and methodsPatient population and specimens analyzedThe study was conducted during a period of one year from April 2012 to March 2013. A total of 150 biological specimens were collected from patients with a clinical diagnosis of EPTB, and 20 negative control biological specimens were collected from patients with an alternative diagnosis. The details of the specimens analyzed are given in Table 1.
Sample processingAll the samples were subjected to Direct Fluorescence staining (DF), mycobacterial culture, and multiplex PCR assay. Patient demographics were collected from the case files as well as with the help of respective clinicians.
The diagnosis of tuberculosis was made on the basis of a CGS which included clinical characteristics, microbiology smear as well as culture, histopathology, cytology, radiology, and response to antitubercular therapy. A case was considered true positive if culture was positive or any two of the other criteria mentioned in the CGS.
DF staining was done using Auramine-Rhodamine (Sigma/Himedia) staining and examined using fluorescent microscope.8 For mycobacterial culture, urine and tissue samples were subjected to digestion and decontamination by N-acetyl-l-cystine NaOH (NALC-NaOH) method.9 Other samples from sterile sites were subjected to decontamination only if a Gram stain or routine bacterial culture showed the presence of any organism, otherwise were processed directly. All the samples were subjected to culture by BacT/ALERT 3D (biomerieux, Durham, North Carolina, USA) and LJ media as per the standard protocol.10
Identification of mycobacteriaPositive growth in either media was identified as M. tuberculosis (MTB) using AccuProbe molecular identification system (GenProbe, San Diego, California).9 AccuProbe culture identification test was performed as described by the manufacturer. For the identification of non-tuberculous mycobacteria (NTM), GenoType Mycobacteria CM (Hain Lifescience GmbH, Nehren, Germany) was used as recommended by the manufacturer.11 The assay permits the simultaneous molecular genetic identification of the M. tuberculosis complex and 14 of the most common NTM species (M. avium ssp., M. chelonae, M. abscessus, M. fortuitum, M. gordonae, M. intracellulare, M. scrofulaceum, M. interjectum, M. kansasii, M. malmoense, M. peregrinum, M. marinum/M. ulcerans, and M. xenopi) from cultivated samples.
Real time multiplex PCRStandardization of PCRValidation of the real time PCR was done using known positive and negative samples as well as the ATCC standard strain H37 Rv spiked samples. For detection of the analytical sensitivity of the assay, H37 Rv strain was subjected to PCR using both targets (IS6110 and MPB64) in different dilutions. 0.5 McFarland standard was prepared and was serially diluted to achieve different concentrations as 108, 107, 106, 105, 104, 103, 102, and 101bacilli/mL of the sample. Multiplex real-time PCR was run using different dilutions of H37 Rv spiked samples, three times using each dilution.
DNA extractionSamples after subjecting to decontamination by NALC/NaOH method, the DNA extraction was done using spin column method using DNASure Tissue Mini Kit (Nucleopore, G Brand, Genetix Biotech Asia Pvt. Ltd., New Delhi, India) as per the manufacturer's instructions. Briefly, 25mg of tissue or 1–2mL of other samples were placed in a microcentrifuge tube. 180μL of lysis buffer and 25μL of proteinase K solutions were added and vortexed. Incubated at 56°C for 1–3h, vortexing in between till complete lysis of the sample was obtained. 200μL of lysis buffer 3 was added, vortexed and incubated at 70°C for 10min. If insoluble particles were visible, centrifuged for 5min at 11,000×g and transferred the supernatant to a fresh microcentrifuge tube. 210μL of absolute ethanol was added and vortexed vigorously. For each sample, one DNASure mini kit column was placed into a collection tube and the sample was applied to the column. Centrifuged for 1min at 11,000×g and discarded the flow-through and placed the column back into the collection tube. Washed using wash buffer twice and discarded the flow-through. Placed the column into 1.5mL microcentrifuge tube and added 100μL prewarmed buffer (70°C), incubated at room temperature for 1min and centrifuged at 11,000×g for 1min. The extracted DNA was stored at −70° C till the test was performed.
PCR protocolAmplification of gene targets was done using Multiplexed Tandem PCR employing two sequential PCR steps automated by the Easy-Plex™ system (AusDiagnostics Ptv. Ltd., Alexandria, New South Wales, Australia) targeting two mycobacterial DNA sequences, IS6110 and MPB 64, and also Nono human reference gene for DNA extraction control and Spike target for PCR inhibitor control. The Step 2 PCR reaction was performed in Rotor-Gene 6000 (Qiagen, Germany) thermal cycler. The product from Step 1 was used for real time PCR reactions in Step 2 using primers ‘nested inside’ the primers used for Step 1, to increase the sensitivity and specificity of the assay, by fundamentally blocking non-specific priming.
Master-Mix reagents and 72-well gene discs containing lyophilized primers were prepared by AusDiagnostics Pvt. Ltd. (Alexandria, New South Wales, Australia). The M. tuberculosis specific IS6110 and MPB 64 gene loci were used to design primers. 10μL of the purified DNA was used directly for PCR. First-round multiplexed preamplification was performed in a 20μL reaction volume. Step 1 is a short (18 cycles) multiplexed pre-amplification reaction using primers homologous to all targets, IS6110, MPB64, Nono; human reference gene and Spike; PCR inhibitor control. 18 cycles were performed, denaturation of ds DNA at 95°C, annealing of primers at 60°C and extension at 72°C. Dilution of PCR amplicons (product from step 1) was done by biorobot in a plate placed in Easyplex system. Diluted amplicons were added in 72-well Easy-Plex™ ring containing primers for individual targets. The amplification of each target individually in separate wells, in the step 2 of PCR, helped us improving the sensitivity and specificity of the assay. This 72 well gene disc was sealed in heat sealer and placed in Rotor gene. Real time amplification was performed on a RotorGene thermal cycler RG6000 using primers ‘nested inside’ the primers used for Step 1. The thermal cycling conditions were 95°C for 10min followed by 30 cycles of 95°C for 15s, 60°C for 15s, and 72°C for 15s. DNA amplification is measured by the increase in fluorescence when Eva-Green™ dye was incorporated into the DNA being formed in the specific amplification reaction.
Presence of either of the sequences was considered as a positive result and a comparison between the two targets was also done. Comparative sensitivity for detection of M. tuberculosis was also done for these two sequences.
Quality controlThe PCRs were monitored for inhibition by the inclusion of an internal positive control (Spike; consisting of artificial DNA) with each specimen. For extraction procedure, human reference gene, Nono was used as the system control. Tests were also monitored for contamination using water as a negative control in each test run. Each PCR run included ATCC standard M. tuberculosis strain H37 Rv as positive control.
Statistical analysisSensitivity, specificity, positive and negative predictive values of the multiplex PCR as well as that of individual targets (IS6110 and MPB64) were calculated against the CGS. The significance of difference was determined by proportion test; probability value of 0.05 was taken as significant value (p<0.05).
Ethics approvalStudy was approved by the hospital ethics committee (reference number EC/11/11/313 and approval letter dated 29/11/2011). Consent was waived since this was an anonymized study on samples submitted to mycobacteriology laboratory for diagnosis of TB.
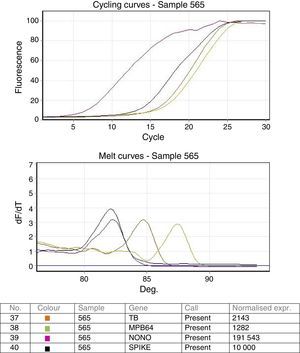
ResultsStandardization of real time PCRValidation of the real time PCR was done using known positive and negative samples as well as the ATCC standard strain H37 Rv spiked samples. Both the targets IS6110 as well as MPB64 were detected in all dilutions in all the 3 repeat runs. Analytical sensitivity of the real time multiplex PCR was found to be as low as 10bacteria/mL of the sample. Cycling and melting curves of the multiplex PCR are shown in Fig. 1.
Smear and culture resultsOut of the total 150 extrapulmonary samples included in the study, two samples failed the extraction of DNA and were excluded from the analysis. 30/148 (20.3%) specimens were positive by DF staining. 35/148 (23.6%) specimens were culture positive for M. tuberculosis. The average time to detection for culture positive samples was 23.6 days
Real time PCR resultsOut of the 148 samples subjected to PCR test, 67 (45.3%) were positive and 81 (54.7%) were negative by PCR. Details are provided in Table 2. Out of the 67 PCR positive samples, 41 were microbiology confirmed, 16 were histopathology/clinical/radiology positive and 10 samples were negative by all other tests. Among 81 PCR negative samples, five were positive using the CGS (four were microbiology positive & one histopathology/clinical/radiology positive), and 74 were negative by all the other tests (Table 2).
Multiplex PCR results in extrapulmonary tuberculosis in comparison to other modalities of diagnosing tuberculosis.
| Sample type | PCR positive, n=67 (45.3%) | PCR Negative, n=81 (54.7%) | ||||
|---|---|---|---|---|---|---|
| Microbiology smear/culture positive | Microbiology negative, histopathology/clinical/radiology positive | All negative | Microbiology smear/cuture positive | Microbiology negative, histopathology/clinical/radiology positive | All neg | |
| Body fluid (n=30) | 4 | 4 | 3 | 1 | 0 | 18 |
| Tissue (n=46) | 12 | 5 | 4 | 1a | 1 | 23 |
| Pus/aspirate (n=61) | 25 | 6 | 3 | 3+1a | 0 | 23b |
| Urine (n=11) | 0 | 1 | 0 | 0 | 0 | 10 |
| Total (n=148) | 41 (61.2%) | 16 (23.9%) | 10(14.9%) | 6 (7.4%) | 1 (1.2%) | 74 (91.4%) |
PCR, polymerase chain reaction; NTM, nontuberculous mycobacteria.
Out of the, six microbiologically positive and PCR negative samples (Table 2), two smear positives subsequently grew NTM and the remaining four samples were only culture positive for MTB. These two samples that grew NTM on culture were treated as microbiologically negative (since they did not grow MTB), while calculating sensitivity and specificity of the PCR. The sensitivity and specificity of the multiplex PCR obtained was 91.9% and 88.4%, respectively, when CGS was used as the gold standard. The positive and negative predictive values of the multiplex PCR (using both the targets) were estimated as 85.1% and 93.8%, respectively. However, when the 20 control samples were used to assess the negative predicative value, it was found to be 100%. When the assay performance was analyzed for different samples, the sensitivity varied from 88.9% for body fluids, 91.2% for pus/aspirate samples to 94.4% for tissue samples. The specificity of the assay varied between 85.2% for tissue, 85.7% for body fluids to 88.5% for pus/aspirate samples. Even though the test was 100% sensitive and specific for urine samples, the sample size was too small for interpretation.
Results of smear and culture in 67 PCR positive samples are given in Table 3. Ten culture negative samples were positive by smear as well as by PCR (Table 3). Six out of these were already on ATT for a period varying from one to five weeks, which might have affected the culture results but appear to have not affected the molecular test.
Comparison of smear & culture in PCR positive samples (n=67).
| Sample type | Only smear positive | Only culture positive | Both smear & culture positive | Both smear & culture negative |
|---|---|---|---|---|
| Body fluid | 3 | 1 | 7 | |
| Tissue | 2 | 6 | 4 | 9 |
| Pus/Aspirate | 8 | 4 | 13 | 9 |
| Urine | 0 | 0 | 0 | 1 |
| Total | 10 (14.9%) | 13 (19.4%) | 18 (26.9%) | 26 (38.8%) |
PCR, polymerase chain reaction.
Out of 148 samples investigated, six samples grew NTM, two smear positive and four smear negative. All these six samples which grew NTM on culture were considered as microbiologically negative samples since the focus of the study was MTB. Among the two smear positives, one tissue sample (lymph node biopsy) grew M. avium and the other pus sample grew M. abscessus. The remaining four smear negative samples, which grew NTM, were pus samples and were identified as M. fortuitum (1) and M. abscessus (3). All these six samples were PCR negative for both targets.
Among the 67 PCR positive samples, 39 were positive for both targets, 27 only for IS6110 and one sample only for MPB64 (Table 4). Out of the 148 samples analyzed, more positive results were observed with target IS6110 (44.6%) as compared to target MPB64 (18.9%). Only one sample out of 148 (0.68%) was positive only for MPB64 and was negative for IS6110. The sensitivities of individual targets IS6110 and MPB64 were 90.3% and 64.5%, respectively, and specificities 88.4% and 97.7%, respectively, in comparison to the CGS. While IS6110 was found to have much higher sensitivity as compared to MPB64 (p<0.001), MPB64 had better specificity (p<0.05).
There were 10 out of 67 PCR positive samples that were negative according to the CGS. The details of these samples are given in Table 4. All 20 negative controls from patients with no clinical suspicion of tuberculosis and with an alternative diagnosis were negative by multiplex PCR using both targets.
DiscussionTB remains a major global public health problem, especially in India. Still worse is the fact that the diagnostic tools to detect EPTB are not sensitive enough to aid in the early detection of such cases. The clinical utility of detecting M. tuberculosis by PCR depends on its rapidity and accuracy in diagnosing EPTB and has been evaluated in the current study. We have analyzed the accuracy of the PCR assay in the diagnosis of EPTB using a CGS and have also analyzed the comparative efficiency of two commonly used PCR targets in India, namely IS6110 and MPB64.
The role of PCR in the diagnosis of EPTB has been evaluated previously as an alternative diagnostic tool and has yielded variable results with sensitivities ranging from 42 to 100% and specificities ranging from 85 to 100% using various PCR targets such as IS6110, MPB64, TRC4, GCRS, etc.12–14 The present multiplex PCR targeting IS6110 and MPB64 seems to be promising with a good sensitivity and specificity of 91.9% and 88.1%, respectively, when CGS was taken as the gold standard. Whereas smear and culture were positive in 47 out of the total 62 cases using the CGS, PCR detected 57/62 positive cases. Thereby 10 additional cases which could have gone undetected by conventional microbiologic methods were detected by PCR. Few recent studies also concluded PCR to be a rapid and sensitive method for diagnosing EPTB compared to conventional microbiologic methods.15,16 In an earlier study by us it was observed that the positivity rate of mycobacterial culture was only 13.6% in extrapulmonary samples in contrast to 18.9% in pulmonary samples.17
However, other 10 doubtful cases were also detected as positive by PCR out of the 84 cases classified as negative using the CGS criteria (Table 5). These were only clinically suspected cases, but none of the other parameters turned out positive, other than PCR which resulted in lower specificity of multiplex PCR to 88.4%. For these cases, it was not possible to ascertain whether these were true cases of tuberculosis. However, all the 20 negative control samples from patients with no risk factors for tuberculosis (true negative controls) were negative by multiplex PCR giving 100% specificity to the assay. Moreover, all six study samples which grew NTM on culture were also negative by PCR.
CGS negative, but PCR positive samples (n=10).
| Sample type (no. of samples) | Significant clinical/laboratory findings |
|---|---|
| Pleural fluid (2) | Pleural effusion |
| CSF (1) | SLE patient on steroids |
| Tissue – peritoneum (1) | Peritoneal adhesions |
| Tissue – pleural (1) | Empyema |
| Tissue – rectal (1) | Rectal fistula, histopathologically: nonspecific acute colitis |
| Tissue – nasal (1) | Histopathologically: Inflammatory nasal polyp |
| Aspirate (1) | Nonspecific arthritis |
| Pus (2) | 1) Pyoneprosis, isolated E. coli on routine bacterial culture 2) On cytopathology of submandibular lymph node revealed sialadenitis with reactive lymphadenopathy. |
PCR, polymerase chain reaction; CGS, composite gold standard.
Among the six PCR negative and microbiologically positive samples, two were smear positive and grew NTM on culture (Table 2). The remaining four samples which grew M. tuberculosis, two samples seemed to have very low bacterial load because the culture came out positive after 38 days and 40 days of incubation. In the remaining two samples, one each of CSF and lymph node aspirate, where cultures came out positive within 20–30 days but PCR remained negative.
In addition to the improvement in the diagnosis of EPTB using PCR, data on comparison of commonly used PCR targets in India is also needed. The results of the present study shows that IS6110 with a sensitivity of 90.3% is a better target for amplification of DNA from extrapulmonary samples compared to MPB64 with a sensitivity of 64.5% (p<0.001). Few previous studies also documented increased positivity of IS6110 target in extrapulmonary samples compared to other commonly used PCR targets.18,19 Negi et al. analyzed various targets specific for M. tuberculosis and reported the highest PCR positivity rates when IS6110 was used, but MPB64 was not a target included in their study.18 In our study, out of the 148 samples analyzed, only one sample was IS6110 negative and MPB64 positive. The absence of IS6110 copies in Indian M. tuberculosis strains, as observed by some earlier studies was not very evident in our study. A study by Singh et al. on lymph node biopsy samples also showed IS6110 (69.1%) as a better single target compared to MPB64 (48.2%).20 In their study, while only IS6110 was positive in 24/81 (29.6%) cases, addition of MPB64 detected 7/81 (8.6%) additional cases. Therese et al. in Tamil Nadu concluded that simultaneous use of both targets, IS6110 and MPB64, together on clinical specimens (mostly respiratory specimens) could attain 100% sensitivity in culture proven cases.21 Addition of MPB64 detected only additional 1.4% of the cases in their study. In a study by Sharma et al. the positivity rates of Ziehl-Neelsen smear, culture, and multiplex PCR were 30%, 26.3%, and 91.3%, respectively, in confirmed tubercular lymphadenitis patients.22 This study showed MPB64 (87.5%) as a superior target than IS6110 (85%) in contrast to our study. In our experience, only 1/148 samples (0.68%) was positive by MPB64 and negative by IS6110. However, MPB64 as a single target had a higher specificity (97.7%) than IS6110 as a single target (88.4%) when CGS was taken as the gold standard (p<0.05).
A recent meta-analysis on Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA, USA) where rpoB gene is the target for the diagnosis of EPTB has come up with a pooled sensitivity of 81.2% for lymph node tissues, 62.8% for CSF, and 21.4% for pleural fluid and a pooled specificity of >98.7% against a composite reference standard.23 Our multiplex PCR targeting IS6110 and MPB64 showed a much higher sensitivity (88.9% for body fluids, 91.2% for pus/aspirate samples, and 94.4% for tissue samples) compared with Xpert MTB/RIF assay while the specificity was lower in our study (85.2% to 88.5%) when CGS was used as the reference.
The present study has made an attempt to use CGS for comparative evaluation of multiplex PCR instead of using only culture positivity as gold standard. The variety of extrapulmonary samples included in our study is also an added advantage in deciding the clinical utility of such assays in various clinical settings. But the low sample size especially when individual sample types were analyzed separately is a limitation of our study.
In summary, the multiplex PCR assay targeting IS6110 and MPB64 has a great potential to improve the rapid diagnosis of extrapulmonary tuberculosis. While multiplex PCR detected M. tuberculosis in less than a day, the average time to detection for automated culture was 23.6 days for extrapulmonary samples in our study. Therefore, the suggested protocol is to use PCR along with the microbiological methods to obtain a rapid and accurate diagnosis of TB.
Conflicts of interestThe authors declare no conflicts of interest.
Authors acknowledge the financial support provided by the research development program, Sir Ganga Ram Hospital.