Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB), is one of the top 10 causes of death worldwide. Drug-resistant tuberculosis (DR-TB) poses a major threat to the World Health Organization's “End TB” strategy which has defined its target as the year 2035. In 2019, there were close to 0.5 million cases of DRTB, of which 78% were resistant to multiple TB drugs. The traditional culture-based drug susceptibility test (DST - the current gold standard) often takes multiple weeks and the necessary laboratory facilities are not readily available in low-income countries. Whole genome sequencing (WGS) technology is rapidly becoming an important tool in clinical and research applications including transmission detection or prediction of DR-TB. For the latter, many tools have recently been developed using curated database(s) of known resistance conferring mutations. However, documenting all the mutations and their effect is a time-taking and a continuous process and therefore Machine Learning (ML) techniques can be useful for predicting the presence of DR-TB based on WGS data. This can pave the way to an earlier detection of drug resistance and consequently more efficient treatment when compared to the traditional DST.
In 2019, there were about 0.5 million cases of drug-resistant tuberculosis (DR-TB), of which 78% were resistant to multiple TB drugs. The problem of multi-drug resistance TB (MDR-TB), defined as simultaneously resistance at least to rifampicin (RR-TB) and isoniazid (INH), the two most effective first-line anti-TB drugs, complicates TB management (i) since it requires longer treatment with drugs that are more expensive and toxic (recommended a treatment regimen that includes second-line drugs for people with MDR/RR-TB), and (ii) by the rising number of incidences of MDR/RR-TB worldwide.1
To interrupt the chain of TB transmission and avoid the development of DR-TB, there is need for rapid diagnosis, application of an adequate treatment regimen and (regular) close follow-up of the patient. To speed up diagnosis and generation of the drug resistance profile, some methods based on nucleic-acid amplification test (NAAT) are already endorsed by WHO such as the Xpert-MTB-RIF ULTRA (Cepheid, Sunnyvale, CA, USA), focusing only on the principal mutations associated with rifampicin-resistance. For the better, WGS based techniques can predict the drug resistance profile revealing known mutations2–6 and can be utilized for proposing new resistance conferring mutations.7,8
In a comparative analysis of various drug resistance profile methods, WGS has been shown to be a reliable technique among the genotypic tests when compared to various DST, being not only accurate, but also providing a rich set of additional information for further analysis.9 Therefore, WGS is the basis of new insight into the genetic basis and unknown mechanisms of drug resistance and as such, is essential for the development of new antibiotics for TB.10 For this reason, WGS is being proposed as the reference technique for detecting mutations associated with DR-TB.11
It is therefore critical to explore the value of modern statistical approaches such as Machine Learning (ML) to assist rapid clinical diagnostics based on the predicted drug resistance profile directly from the WGS data derived from DNA, extracted from MTB cultures,4,12–16 as well as clinical specimens such as (mostly) sputum.3,17,18
Recently, there have been many studies which explored various classes of algorithms for predicting the drug resistance profile from WGS data. One of the earliest included a statistical and rule based (Direct Association) approach19,20 that helped to establish the feasibility of relying on WGS data as the basis for further analysis, compared to the DST. The dataset, techniques and limitations discussed in these papers19,20 have been studied and analysed further in the last five years.7,8,14,21,22 The drug resistance profile of a sputum sample can be predicted within five days after sampling which is roughly 24 days earlier than the WGS from Mycobacterial Growth Indicator Tube (MGIT) culture, and up to 31 days earlier than DST; in addition, WGS-based DR prediction is less expensive.23 There is strong evidence favouring the hypothesis that direct WGS on sputum combined with specialized software tools shall allow almost real-time diagnosis and surveillance ability in a cost-effective manner,24 when compared to the traditional approaches.
Despite the growing number of studies presenting and evaluating tools for assessing drug resistance profile based on WGS data, the concepts and application are not widely discussed within the biomedical environment and ML is perhaps still relatively underutilized in clinical applications. Thus, in this study we aimed to (i) describe and discuss the main approaches for WGS analysis in TB diagnosis and detection of DR, and (ii) analyze the TB drug resistance profiling based on ML through a literature review.
MethodologyThe data was collected through an in-depth search of the various publications in PubMed as well as WHO publications including the words “whole genome sequencing” AND/OR “drug resistance prediction”, AND/OR “machine learning”, AND/OR “tuberculosis incidence surveys”, AND/OR “genomic medicine”, AND/OR “clinical application of machine learning algorithms” and analyzed based on the criteria of direct relevance to the problem of drug resistance profiling using ML on WGS data of MTB.
The inclusion criteria were: (i) the article having been published no longer than six years ago and (ii) the sample size being larger than 500 genomes. The exclusion criteria were: (i) the absence of supplementary material such as sample IDs from NCBI and, (ii) the study being limited to the use of classical statistical techniques.
Results and discussionDrug resistance prediction using direct associationConventional WGS based drug resistance prediction methods rely on identification of the number and nature of mutations, such as (mostly) Single Nucleotide Polymorphism (SNP) and Insertion-Deletion (INDELS) as compared to a reference genome, together with correlation with drug resistance profile obtained by conventional DST. This method is driven by a pre-documented library of resistance conferring SNP and is therefore called as Direct Association method.6,19,20
The knowledge of resistance conferring SNP and its correlation with drug resistance profile is a meticulous process which demands extensive experimentation and literature review to confirm the nature and location of the mutations.6,25–29 An incomplete understanding of these mutations and their effects, whether directly or indirectly, limits the accuracy of molecular diagnostic tools.
Various online databases have been developed to provide centralized resources to identify the mutations and to predict their effects, including (i) MycoResistance,30 (ii) Tb-Portals,31 (iii) TBDReaMDB,32 (iv) Mubii-TB-DB,33 (v) ReSeqTB34 and most recently by WHO.6
A study by Walker et al.19 applied WGS on 2,099 MTB isolates to identify and classify common, as well as rare mutations, which predict drug resistance profile of a particular MTB sample for first and second-line drugs. This was achieved by devising a classification algorithm for the observed resistance-conferring mutations in the isolates, that was compared to the reference genome to the SNP both in coding sequences (CDS) and their promoter regions as well as the presence (insertion) and absence (deletion) of amino acids in the sequence.
Another study by Allix-Béguec et al.20 evaluated the hypothesis that if all the resistance-conferring mutations were known extensively, it should be possible to infer the drug resistance profile from the presence or absence of these mutations since the WGS data provides information for virtually all the genomic sites of interest associated with drug resistance. The conclusions were favourable to the use of WGS data, but the study also highlighted that, the drug resistance profile interpretation might be complicated by the underlying biological processes (e.g., gene-gene interaction) which are still underexplored. The WHO has also published a condensed catalogue of confidence-graded mutations that have established correlations with DR in MTB6 that enumerates all the mutations showing statistically significant experimental association to resistance.
Software such as TB-profiler,35,36 Kvarq37 and MTBSeq38 are based on algorithms relying on Direct Association to predict an isolate's drug resistance profile from their particular SNPs.
Recently, specifically for MDR, advanced molecular tests have been developed, using the direct association approach as well, but these also suffer from drawbacks such as inability to model resistance conferring gene-gene interactions. In addition, these tests are based on selective amplification of antibiotic resistance associated genes for only a subset of drugs used for TB treatment and therefore are less predictive for drugs such as the second and third line of drugs.8
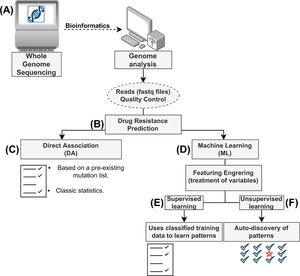
Drug resistance prediction using machine learningML is a field of computer science which aims to utilize available data to discover patterns, infer knowledge and then make decisions based on this knowledge towards similar yet unseen data. ML is further divided into (i) supervised learning and (ii) unsupervised learning (Fig. 1).
Flow diagram for prediction of drug resistance from whole genome sequencing (WGS) data using computational approaches. (A) The data generated from WGS (FASTQ files) for (B) predicting drug resistance either using (C) the classical Direct Association, which relies on a database of documented mutations at present or (D) Machine learning techniques, such as (E) Supervised Learning, which relies on guided training of algorithms on hand-curated data to predict the effects of novel mutations or (F) Unsupervised Learning, which relies on algorithmic techniques to discover patterns and predict effects of the mutations.
Supervised learning is the most used category of ML algorithms, that makes use of labelled dataset (training data) and generates a generalized model which is used to make predictions about unseen data. Some examples of supervised learning algorithms are Linear Regression, Logistic Regression, Support Vector Machine.
Unsupervised learning, on the other hand, uses unlabelled data as a training dataset and then tries to generalize the inference or prediction model.39 Some examples of unsupervised learning algorithms are Neural Networks, Principal Component Analysis and K-means clustering.
Deep learning, another branch of ML has been successfully applied in multiple fields40 and is rapidly gaining popularity in computational biology. This success is however, not without some reservations, since deep learning effectively relies on some very advanced mathematical concepts which are difficult to discern.41 One crucial aspect in computational approaches is the identification and refinement of important variables (called features) for the application of theses algorithms.42
The biological processes which result in mutations giving rise to more complex gene-gene interaction and therefore lend themselves quite naturally to multi-variate analysis techniques and can be better explored through ML algorithms.
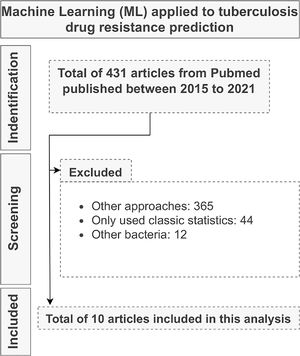
Machine learning applicationAfter examining the title, abstracts, and conclusions of the initially considered 431 papers, a total of 10 papers were selected for full-text analysis (Fig. 2). For the 10 studies, we highlighted (i) the algorithms used, (ii) the applied evaluation metrics, (iii) the sample size, and (iv) their main conclusions (Supplementary table 1).
The study by Yang et al.14 investigated the performance of multiple classification algorithms with eight drugs as target labels for DST prediction. The predictions were based on the data generated by feature engineering on 23 candidate genes and their 100 base-pair upstream regions,19 and were then validated against the DST results for all 1,839 samples leading to a total of 2,629 SNPs (dimensions), that were analyzed in three feature sets of SNPs, with the assumption that all polymorphisms are resistance determining.
The authors also explored clustering techniques such as Principal Component Analysis and Sparse Logistic Principal Component Analysis to reduce the number of dimensions from 2,629 to two as principal components. These principal components were used for a cluster analysis and with the observation that Sparse Logistic Principal Component Analysis performed better in identification of distinguished classes within a cluster.
The authors reported that the best performing models, when compared to the rules based on Direct Association, performed favourably with an increase in sensitivity for these eight drugs. Models generated using the Product-of-Marginals and the Support Vector Machine with Radial Basis Function kernel were the best performers. These algorithms improved the mean sensitivity in terms of resistance classification as well as the area under the ROC curve (AUC) for first line of drugs and the prediction results were comparable to the results with respect to Direct Association based drug resistance profiling.
In a follow up study by Yang et al. 43, the authors evaluated a Multi-task with Deep Denoising Auto-encoder algorithm, that simultaneously classifies an isolate based on the resistance profile, against four drugs on a cohort of 8,388 MTB isolates. Furthermore, the models were evaluated on the genomic data using the same pre-processing techniques used in their earlier study.44 The authors noted that for MDR-TB, the Deep Denoising Auto-coder algorithm, that relies on non-linear dimensionality reduction suitable for sparse datasets (like mutation datasets) performed better than other algorithms in the study, while achieving best sensitivity scores against all drugs, except for rifampicin. In conclusion, the authors noted that the traditional ML performance metrics might not be suitable for the comparison and validation of multi-task algorithms for clinical application.
Another study by Chen et al.8 explored deep learning models to predict the drug resistance profile against multiple TB drugs based on the analysis of SNPs and INDELs, using a novel Multi-task Wide and Deep Neural Network, that was evaluated on a cohort of 3,601 MTB strains containing 1,228 MDR-TB strains, against 11 drugs. The initial set of features identified by the authors consisted of a total of 6,342 different INDELs and SNPs in 30 promoter, intergenic, and coding regions, which were then reduced, by considering their presence across the cohort, using feature engineering techniques for aggregating and deriving a final set of 222 features.
`The authors described the novel features of Multi-task Wide and Deep Neural Network as a combination of two models Logistic Regression, the “wide” aspect of the neural network being an advantage while modelling the effect of individual mutations and Multi-layer Perceptron model, the “deep” aspect of the neural network being an advantage allowing for modelling the complex epistatic effects to influence the predictions.
The models considered in the study were, Single-task Wide and Deep Neural Network (trained for all drugs individually), Multi-task Wide and Deep Neural Network, Random Forest, Logistic Regression were trained on a full set of 222 features, except for Multi-layer Perceptron, which was trained only upon drug-specific resistance conferring features.
The authors highlighted that there were significant performance gains when compared to regularized Logistic Regression and Random Forest. It is worth noting that Multi-task Wide and Deep Neural Network was also evaluated on samples that had only been partially phenotyped and the proposed Multi-task architecture shared information across different TB drugs. This was achieved by building upon the inter-drug similarities in resistance pathway information and genotype-phenotype relationship leading to a more accurate phenotypic prediction. The cohort was also analysed based on the 33 lineage defining mutations, to calculate isolate-isolate (pairwise) Euclidean distances ranging from 0 to 3.87, demonstrating five well-defined clusters.
Furthermore, both Single-task (model trained on each drug individually) and Multi-task Wide and Deep Neural Network models were evaluated. In contrast to Single-task models, the proposed Multi-task model predicted the drug resistance profile for multiple drugs, allowing the model to have a holistic view of the resistance pathway information concerning various drugs and while taking into consideration the fact that drug resistance can be caused by both direct genotype-phenotype relationships as well as epistatic effects.
The authors concluded that the Multi-task Wide and Deep Neural Network (i) performed favourably and achieved a higher sum of specificity and sensitivity when compared to all the other models, (ii) was able to rank the mutations according to their confidence level for prediction of resistance, and (iii) was able to share the information / learning across the various drugs rather than treating each drug individually.
In a follow-up study, Chen et al.13 conducted a comparative analysis on the same dataset and the same cohort of algorithms that was used in the previous work8 with the objective to evaluate the utility of training models based on frequent resistance-conferring variants as well as rare variants that are known to be determinants of resistance for at least one drug. The authors did not rely on reducing the number of contributing variables (dimensionality reduction) but rather relied upon an interpretable set of input predictors for Wide and Deep Neural Network and still observed improved performance. This study pinpoints the possibility to predict drug resistance especially for second and third line of drugs, and in particular pyrazinamide, where the individual rare mutations have been shown to be causative.
Other classes of algorithms, known for their interpretability and high accuracy are Classification Trees and Gradient Boosted Trees, that have also been utilized by Deelder et al.7 in a study of 16,688 samples covering four main TB lineages, to uncover novel putative mutations associated with resistance to 14 drugs. The authors leveraged the interpretability of Classification Trees and superior prediction ability of Gradient Boosted Trees for improving the overall resistance prediction as they make fewer assumptions on the distribution and functional relationships between features. It is worth highlighting that in addition to yielding predictions, these algorithms also rank the importance of the features.
These algorithms were trained on various features sets consisting of (i) SNPs in resistance associated genes and (ii) genome wide SNPs, with the inclusion and exclusion of co-occurrent resistance markers. The authors noted that the inclusion of co-occurrent resistance markers for multiple drugs, led to superior results for Gradient Boosted Tree algorithm, in terms of predictive accuracy and of AUC when compared to other models for certain drugs.
However, the authors cautioned against clinical application of the procedure because of the inclusion of co-occurrent resistance markers and the ability of genome-wide Gradient Boosted Tree model to capture covariate interactions, which might not translate optimally into clinical environments, since the markers by themselves, might be more indicative of transmissibility rather than drug resistance. In conclusion, the authors highlighted that the quantitative minimum inhibitory concentration (MIC) scores as phenotypes could be included in a future study to improve the overall prediction metrics.
The study by Kouchaki et al.12 aimed at a large and diverse cohort of 13,402 MTB isolates from multiple MTB lineages across six continents and, including 11 TB drugs, targeting the 23 resistance-conferring genes as established in a study by Walker et al.19 With the increased size of the cohort, the authors aimed to create ML models which were more generalizable (applicable on any general dataset for MTB). The authors observed that with increased size of the dataset, the multi-variate genomic information, grew sparser and the mutation information was spread throughout these numerous variables. Therefore, the effects of dimensionality reduction, through the application of the Sparse Principal Component Analysis / Non-Negative Matrix Factorization algorithms, were also evaluated by the authors. Furthermore, the features were divided into three distinct sets following the work done by Yang et al.44
Algorithms such as Support Vector Machine, Linear Regression and Product-of-Marginals methods were evaluated on the feature space after dimensionality reduction, along with the evaluation of ensemble techniques such as Random Forest, Adaboost and Gradient Boosted Trees. The ML algorithms were able to rank according to importance, known resistance conferring mutations for well-studied first line drugs and also indicated correlation between lineage defining mutations with drug resistance, for the second line drugs.
The authors reported enhancement in the performance of Logistic Regression model and Gradient Boosting Trees model when used in conjunction with a dimensionality reduction of number of features using Sparse Principal Component Analysis / Non-Negative Matrix Factorization algorithms in terms of F1 score (a metric for measuring algorithm's classification power), for second line TB drugs.
In a follow up study, Kouchaki et al. 21, investigated the use of Single-label and Multi-label Random Forest on the same dataset used in the previous study,12 with the dual goal of (i) evaluating its performance on drug resistance prediction and, (ii) ranking mutations by the order of importance. The authors noted that some mutations are commonly identified in MDR-TB and extensively drug resistant (XDR-TB) isolates and suggested that predicting the global phenotype (MDR-TB) rather than the individual phenotype (RR-TB) could be a promising approach. The Multi-label Random Forest algorithm simultaneously classifies the MTB isolate as being resistant to multiple drugs, similar to the Wide and Deep Neural Network algorithm by Chen et al,8 capturing the correlation between the drugs for resistance co-occurrence.
These Single-label and Multi-label Random Forest algorithms were used to target the 23 genes identified by Walker et al,19 observing a total of 5,919 baseline variants from the 23 candidate genes. Due to fewer number of MDR-TB and XDR-TB samples in the cohort, the dataset was treated with a stratified sampling technique to minimize the imbalance in the dataset. Furthermore, five feature sets were created using various sub-sets of mutations, that were used to evaluate the impact of feature sets on the classification performance of the algorithms.
As a sub-study, the authors also trained ML models using only the top-ranked mutations (top 16-37 mutations) to evaluate the overall classification performance of the models and observed favourable results and noting that increasing the number of features improved sensitivity, but reduced specificity. In conclusion, the authors mentioned that the Multi-label Random Forest algorithm had higher sensitivity and lower specificity when trained upon overlapping set of variables (feature sets) derived from the baseline features.
Protein sequences, complementary to genome sequences, have also been explored as a foundation of drug resistance profile by Chowdhury et al. 16, where the authors applied Stacked Ensemble algorithm on features derived from physic-chemical, evolutionary, and structural properties (features) to predict resistance against capreomycin. The algorithms considered in the study were Generalized Linear Model, C5.0, Support Vector Machine and Stacked Ensemble, which combined the other algorithms together into a single classifier.
The authors highlighted that using protein sequences as a foundation offers a better basis for further downstream analysis, since a greater number of features are discernible from protein sequences and relied on the Pearson's correlation coefficient for dimensionality reduction, resulting in the reduction of features from 621 down to 392. The Linear Regression, C5.0 and Support Vector Machine algorithms were used as the base learners in the study, resulting in an overall increase in the performance of the final stacked ensemble model, which combined these base learners, while by itself Support Vector Machine algorithm was identified as a strong performer on the dataset.
For Stacked Ensemble’s meta-learner classifier algorithm, the Generalized Linear Model, Linear Discriminate Analysis and Random Forest were evaluated on their ability to combine the base learners in an effective manner. The stacked ensemble model with the Generalized Linear Model meta-classifier was observed to give the best performance.16 In conclusion, the authors highlighted that protein sequences, as opposed to genome sequences, provide a richer feature set for ML algorithms.
Other factors such as structural information (3-D structural mutation mapping), geographic diversity and pan-genome analysis has also been used as a basis for ML analysis in a study by Kavvas et al. 45. The authors selected a genetically, geographically, and phenotypically diverse MTB cohort of 1,595 strains and analysed them for resistance against 13 TB drugs. Instead of relying upon the alignment-based feature engineering, the authors relied upon allele-based pan-genomic basis for feature engineering which does not reduce non-H37Rv variants to a collection of SNPs, while capturing the strain-to-strain variation observed in the bacterial genomes without biasing the variations relative to a single reference genome such as that of MTB.
Furthermore, to validate the allele-based foundation, the authors utilized statistical metrics such as (i) mutual information and (ii) chi-squared and ANOVA F-test, for the identification of resistance-determining genes with newly constructed variant pan-genome, achieving comparable results to the k-mer based approaches. The feature set derived from alleles was used as a basis for Support Vector Algorithm, due to its ability to utilize the relationships between the features. In conclusion, Kavvas et al.45 outlined that while the algorithm provided insights, into the anti-microbial resistance gene identification process, such as the magnitude and sign of the alleles that represents the allele's contribution to drug resistance; it does not provide any insights into the correlation of any mutations in a specific region with the resistance profile against specific drugs.
Most of the studies have relied on complete (comprehensive) genome sequences, which is ideal although not always possible. Alternatively, Nguyen et al.46 have explored the possibility of predicting the drug resistance profile of a sample after the removal of resistance-conferring genes and focusing only on randomly selected core-genes (25-500), defined as the genes common to members of the same species which are not known for conferring resistance. This exclusion of non-core-genes reduces the chances of considering genes transferred horizontally across species, for training ML models to be evaluated across four different bacterial species.
Multiple XGBoost and Random Forest models47 were trained upon feature sets derived from random selection of core genes ranging from 25 to 500, with similar trends across various species for classification score as well as error rates. The authors noted that (i) the clade size and the distribution of phenotypes within the clades had little effect on the accuracy of the models, (ii) high-importance genes were distributed over the genome of the organism, confirmed by the evolution of models on 100 randomly sampled non-overlapping core gene sets, (iii) the results of the models were not influenced by the choice of algorithm and (iv) the models do not suffer from overfitting, memorization, strain-specific SNPs or sampling imbalances.
ConclusionsThe knowledge and understanding of the biological phenomena behind drug resistance mechanisms and the drug-target interaction is still incomplete. Therefore, the way that ML algorithms model this phenomenon might differ based on the class of ML algorithm selected for a particular drug. There is a stark difference between the availability of the genomic data and analysis (within one to nine days) versus the traditional three to eight weeks in the case of the reference phenotypic evaluation method on which the drug regimen of the treatment is adapted. Once the WGS data of the MTB isolate is made available, it is possible, among other purposes, to predict drug resistance profile based on ML, which can assist the clinical decisions for effective treatment of TB by choosing adequate drugs early on and thereby reducing the risks of generating more difficult to treat forms of TB.
During our literature survey we observed that the current state of knowledge of mutations that, either directly or indirectly, impart drug resistance characteristics to the MTB against specific drug candidates, is not exhaustive. Moreover, there are multiple techniques which have been used for feature engineering ranging from SNPs and INDELs to alleles as well as proteomics. As a result, various classes of ML algorithms have been successful in specific aspects of drug resistance prediction for example against a particular drug, mutation-ranking etc. However, these alone are not indicative of the overall predictive accuracy when we think in terms of clinical applicability.
In the literature survey we observed that the performance and clinical applicability of ML algorithms shows huge variations owing to factors such as (i) sampling of the training data for MTB lineages, (ii) frequency of MDR and XDR samples in the cohort, (iii) differences in feature engineering techniques, (iv) multi-drug prediction accuracy, (v) performance evaluation metrics, (vi) inclusion of non-WGS data in the training dataset such as patient's medical history and (vii) interpretability of the ML models.
It is therefore worth investigating approaches that combine (these) different classification models and help overcome their individual weaknesses, counter misclassification and are adaptable towards the inclusion of other aspects involved in the clinical application. One promising class of algorithm is Stacked ensembles that could combine base learners into an ensemble and has shown promise in its ability to counter each base learner's weaknesses and be more accurate overall as a result. These algorithms were used in the study by Chowdhury et al16 and combined the “base learners” using another algorithm called a “meta-learner” to achieve a better performance and made the model less prone to variability across various drugs.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statementAbhinav Sharma: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Edson Machado: Writing – review & editing, Visualization. Karla Valeria Batista Lima: Supervision, Visualization, Writing – review & editing. Philip Noel Suffys: Supervision, Visualization, Writing – review & editing. Emilyn Costa Conceição: Conceptualization, Methodology, Investigation, Supervision, Writing – review & editing.
We are grateful to Saumya Shah (UpGrad) and Felicia Wells (Stellenbosch University) for technical support.