Epidemiological and molecular data on community acquired methicillin resistant Staphylococcus aureus (CA-MRSA) are still scarce in both Egypt and Saudi Arabia. There is almost no data regarding methicillin resistant Staphylococcus aureus (MRSA) prevalence in both countries. This study was conducted to investigate the prevalence and molecular epidemiology of S. aureus and MRSA nasal carriage among outpatients attending primary health care centers in two big cities in both countries. A total of 206 nasal swabs were obtained, 103 swabs from each country. S. aureus isolates were characterized by antibiotic susceptibility, presence of mecA and PVL genes, SCCmec-typing and spa typing, the corresponding Multi locus sequence typing clonal complex was assigned for each spa type based on Ridom StaphType database. MRSA was detected in 32% of the Egyptian outpatients while it was found in 25% of the Saudi Arabian outpatients. All MRSA isolates belonged to SCCmec type V and IVa, where some isolates in Saudi Arabia remained nontypeable. Surprisingly PVL+ isolates were low in frequency: 15% of MRSA Egyptian isolates and 12% of MRSA isolates in Saudi Arabia. Two novel spa types were detected t11839 in Egypt, and t11841 in Saudi Arabia. We found 8 spa types among 20 isolates from Egypt, and 12 spa types out of 15 isolates from Saudi Arabia. Only two spa types t008 and t223 coexisted in both countries. Four clonal complexes (CC5, CC8, CC22, and CC80) were identified in both Egypt and Saudi Arabia. However, the data collected lacked a representation of isolates from different parts of each country as only one health center from each country was included, it still partially illustrates the CA-MRSA situation in both countries. In conclusion a set of control measures is required to prevent further increase in MRSA prevalence.
Staphylococcus aureus is one of the most isolated bacterial pathogens in humans, while it is the causative agent of a wide panel of infections ranging from superficial lesions to life-threatening septicemia. Over the past decades it has remained among the top six clinically important pathogens.1,2 Methicillin resistant S. aureus (MRSA) has been recognized as the principal health-care associated pathogen (HA-MRSA) worldwide. In the last two decades, community acquired methicillin resistant S. aureus (CA-MRSA) infections have emerged as well.3,4 Moreover, CA-MRSA strains have been reported to be more virulent than HA-MRSA strains.5 Carriage of S. aureus by humans is a natural phenomenon associated with health and disease. Although multiple body sites can be colonized in human beings, the anterior nares are the most frequent carriage sites for S. aureus. It colonizes the nasal mucosa of approximately 30–50% of individuals. Nasal carriage of S. aureus is an important risk factor for a wide range of staphylococcal infections. Accordingly, it has been widely used as an indicator to assess antibiotic resistance of S. aureus and MRSA in different populations.6,7
Molecular typing of MRSA is an important tool for epidemiological surveillance and for development of infection control measures aimed at preventing the occurrence and dissemination of epidemic clones within hospitals, from the community to hospitals, as well as within community. Several methods have been used for S. aureus typing. Spa typing is a reliable, accurate and discriminatory typing method of MRSA; it is based on DNA-sequencing of the repeat region of the Staphylococcus protein A gene (spa), where repeats are assigned a numerical code and the spa-type is deduced from the order of specific repeats. The concordance between spa typing and multilocus sequence typing (MLST) has been confirmed, and consequently spa typing could be used to predict multilocus sequence typing clonal complexes (MLST CCs) defined by eBURST software.8 On the other hand, the feasibility of spa typing as a more expedite and less technically demanding alternative typing method for MRSA has been demonstrated in Canada based on the observed concordance of spa types with pulsed-field electrophoresis (PFGE) for Canadian types of epidemic MRSA.9 Moreover, Vincent et al. (2013) reported on the cost-effectiveness of spa, Staphylococcus chromosomal cassette (SCCmec), and Panton-Valentine leukocidin (PVL) genotyping of MRSA in relation to PFGE and MLST.10
Since its first description, infections caused by CA-MRSA strains have been reported worldwide.11,12 In both Egypt and Saudi Arabia, data on carriage, prevalence and genotyping of CA-MRSA are still scarce.13,14 The objective of this study was to characterize the MRSA clones dissemination among outpatients attending primary health care centers in both countries. In order to characterize the MRSA strains, different molecular typing methods were used including SCCmec typing, spa-typing and the corresponding multilocus sequence typing (MLST) and clonal complex based on the spa types in the Ridom StaphType database.
Materials and methodsStudy area and study populationA cross-sectional descriptive study was conducted from July to October 2011 on a community sample from Al-kanater Alkhyria city, Egypt and Buraydah city, Saudi Arabia. Al-Kanater Alkhyria is one of the major cities in Al-Qalyubia governorate, located about 22km north of Cairo, the capital city of Egypt. Buraydah city is the capital city of AL-Qassim district, located 350km north of Riyadh, the capital city of Saudi Arabia. We randomly collected nasal swabs from 206 outpatients, 103 from each city. All swabs were taken from outpatients attending primary health care centers which are staffed by a group of general practitioners and nurses, delivering services that include family practice, pediatrics, women's care, family planning and dental care. All outpatients included had no history of MRSA infection, hospital admission or nursing home during the previous year, surgery, dialysis, permanent indwelling catheters, or medical devices inserted through the skin. The objectives of the study were explained for the participants, who have accepted to take nasal swabs. For Egyptian outpatients (EGOs), there were 69% females and 31% males, age ranges from six months to 59 years. For Saudi Arabian outpatients (SAOs), 70% were females and 30% males, age ranges from six months to 60 years.
Specimen collection and screening for nasal carriageSpecimens were collected from both anterior nares using sterile cotton swabs moistened with sterile solution of normal saline. All swabs were kept at 4°C for 24h until processing in the laboratory. Each swab was inoculated in tryptic soya broth (Oxoid) and incubated overnight at 37°C to increase the isolation rate of S. aureus. The broth was subcultured on mannitol salt agar (Oxoid) plates and incubated aerobically at 37°C for 48h. All presumptive S. aureus colonies were identified based on colony morphology, Gram staining, production of catalase, tube coagulase, and DNase test. All S. aureus isolates were preserved in glycerol and stored at −80°C for further experiment.
Antimicrobial susceptibility testingAll S. aureus isolates were subjected to antimicrobial susceptibility testing by the standard agar disk diffusion methodology according to Clinical and Laboratory Standards Institute (CLSI). The following panel of antibiotics were used: oxacillin, cefoxitin, vancomycin, erythromycin, clindamycin, gentamycin, trimethoprim-sulphamethoxazole, chloramphenicol, ciprofloxacin, tobramycin, moxifloxacin, and rifampicin. All oxacillin and cefoxitin resistant isolates were further confirmed as methicillin resistant by the ability of the isolates to grow on Muller–Hinton agar supplemented with 4% sodium chloride and 6μg/mL oxacillin.15,16
Molecular characterizationBoth methicillin resistant and susceptible Staphylococcus aureus isolates were grown in tryptic soya broth at 37°C overnight, bacterial cells were harvested in 2.0mL Eppendorf tubes by centrifugation at 10,000–15,000×g for 10–15min, genomic DNA was extracted using miniprep extraction method which was previously described by Wilson.17 The extracted DNA had kept under (−20°C) for further experiments.
All S. aureus isolates were confirmed by PCR amplification of the nuclease gene (nuc gene) using primers previously described.18 Then, all suspected methicillin resistant and susceptible Staphylococcus aureus isolates were examined by PCR for the presence of mecA (methicillin resistance) and PVL (Panton-Valentine Leukocidin) genes using the protocol previously described.19 The SCCmec typing was performed for all MRSA isolates using a multiplex PCR assay described previously by Zhang et al.20
Spa typing was performed for randomly selected 20 MRSA isolates from Egypt, and 15 MRSA isolates from Saudi Arabia. PCR amplification was performed as described previously. The PCR product was purified using the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany) and sequenced with the Big Dye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems Warrington, UK). The sequencing products were purified with a DyeEx 2.0 Spin Kit (QIAGEN) and then prepared for running on an ABI 3100 Avant Genetic Analyzer in accordance with the manufacturer's (Applied Biosystems) instructions.21,22 The sequences were analyzed and A phylogenetic analysis of spa sequences was performed where A Neighbor-joining tree was constructed from spa gene sequences using the BioNumerics software version 7.1 (APPLIED MATHS, Austin, USA). The concordance between spa typing and MLST was evaluated previously. Faria et al.8 have concluded that spa typing could be used to predict MLST CCs defined by eBURST. The corresponding MLST and MLST CCs was assigned for each spa type based on the spa types in the Ridom StaphType database (http://spa.ridom.de/spatypes.shtml) and on the study by Monecke et al., 2011.23
ResultsAntimicrobial susceptibilityAll S. aureus isolates from both Egypt and Saudi Arabia were susceptible to ciprofloxacin, tobramycin, and moxifloxacin. Among MRSA isolates from EGOs 55% showed resistance to erythromycin, 48% to chloramphenicol, 48% to trimethoprim-sulfamethoxazole, 33% to clindamycin, 33% to gentamycin, 9% to vancomycin. Among MSSA isolates 57% were susceptible to all antibiotics used in this study, while 24% showed resistance to erythromycin, 24% to trimethoprim-sulfamethoxazole, 19% to gentamycin, 19% to chloramphenicol (Table 1). Among MRSA isolates from SAOs 58% showed resistance to erythromycin, 50% to chloramphenicol, 46% to trimethoprim-sulfamethoxazole, 11% to clindamycin, 27% to gentamycin, 4% to vancomycin. Among MSSA isolates 63% were susceptible to all used antibiotics, while 25% showed resistance to erythromycin, 25% to trimethoprim-sulfamethoxazole, 13% to gentamycin, 13% to chloramphenicol (Table 1).
Antimicrobial susceptibility profile of MRSA and MSSA isolates in Egypt and Saudi Arabia.
| Profile | OX | FOX | E | CM | GM | VA | C | SXT | RD | CIP | TOB | MXF | Egyptian isolates | Saudi Arabian isolates |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | R | R | S | S | S | S | S | S | S | S | S | S | 9 | 8 |
| II | R | R | R | S | S | S | R | R | S | S | S | S | 7 | 9 |
| III | R | R | S | R | S | S | R | R | S | S | S | S | 2 | 0 |
| IV | R | R | S | S | R | S | S | S | S | S | S | S | 2 | 3 |
| V | R | R | R | S | R | S | R | S | S | S | S | S | 4 | 3 |
| VI | R | R | R | R | S | S | S | R | S | S | S | S | 4 | 2 |
| VII | R | R | S | R | R | S | S | S | S | S | S | S | 2 | 0 |
| VIII | R | R | R | R | R | R | R | R | R | S | S | S | 3 | 1 |
| IX | S | S | S | S | S | S | S | S | S | S | S | S | 12 | 15 |
| X | S | S | S | S | R | S | R | S | S | S | S | S | 4 | 3 |
| XI | S | S | R | S | S | S | S | R | S | S | S | S | 5 | 6 |
R, resistant; S, susceptible; OX, oxacillin; FOX, cefoxitin; E, erythromycin; CM, clindamycin; GM, gentamycin; VA, vancomycin; C, chloramphenicol; SXT, trimethoprim-sulfamethoxazole; RD, rifampicin; CIP, ciprofloxacin; TOB, tobramycin; MXF, moxifloxacin.
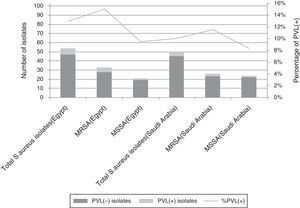
All S. aureus isolates were further confirmed by the presence of nuclease gene. Presence of mecA gene was the final marker for confirming MRSA isolates. Out of 103 screened EGOs, S. aureus nasal carriers were detected in 54 (52%) being MRSA in 33 carriers (61% of S. aureus carriers and 32% of all isolates). For SAOs, 50/103 (48.5%) were S. aureus carriers being MRSA in 26 outpatients (52% of S. aureus carriers and 25% of all isolates) (Fig. 1).
Presence of PVL geneOut of 33 MRSA isolates from EGOs, 5 (15%) were PVL positive, and out of 21 MSSA isolates 2 (9.5%) were PVL positive. Out of 26 MRSA isolates from SAOs, 3 (11.5%) were PVL positive, and out of 24 MSSA isolates 2 (8%) were PVL positive (Fig. 1).
SCCmec typingAmong Egyptian MRSA isolates there were 17 (51.5%) isolates belonging to SCCmec type V, and 16 (48.5%) to SCCmec type IVa. Among Saudi Arabian MRSA isolates there were 11 (42%) isolates belonging to SCCmec type V, and 6 (23%) to SCCmec type IVa, while 9 (35%) isolates remained nontypeable.
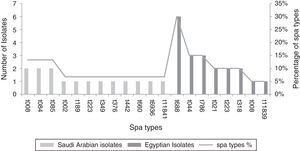
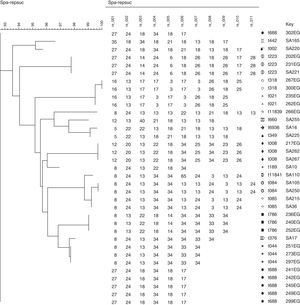
Spa typingSpa typing was performed in randomly selected 20 isolates (60.6% of all MRSA isolates) from Egypt, and in 15 isolates (57.6% of all MRSA isolates) from Saudi Arabia. We found eight different spa types among the 20 Egyptian MRSA spa sequences, and 12 different spa types out of the 15 Saudi Arabian MRSA isolates spa sequences (Fig. 2). Two novel spa types t11839 in Egypt and t11841 in Saudi Arabia were detected and reported to RIDOM spa server. Only two spa types t008 and t223 coexisted in both countries with high frequency of t008 in Saudi Arabia but low frequency in Egypt, all other spa types were different. Six more spa types were identified in Egypt, while 10 more spa types were observed in Saudi Arabia (Fig. 2).
Corresponding clonal complexThe Egyptian MRSA isolates were associated with six clonal complexes including CC5 (t688), CC8 (t008), CC22 (t223), CC30 (t021, t318), CC80 (t044), CC88 (t786), while one spa type was not assigned while it was a novel type (t11839). Among MRSA from Saudi Arabia, the isolates were assigned to seven clonal complexes including CC5 (t002), CC8 (t008), CC15 (t084, t084), CC22 (t223), CC25 (t349, t660), CC80 (t376), CC188 (t189). In addition, three spa types were not yet assigned, and one of them was a novel spa type. Four clonal complexes (CC5, CC8, CC22, and CC80) were identified in both Egypt and Saudi Arabia; other clonal complexes were different (Table 2).
Molecular characters of randomly selected MRSA Isolates in Egypt and Saudi Arabia.
| Isolates | MLST CCs (incidence rate%) | Spa type | No. of Isolates | SCCmec | PVL |
|---|---|---|---|---|---|
| Egyptian spa typed isolates | CC5 (30) | t688 | 6 | V | Neg |
| CC8(5) | t008 | 1 | V | Pos | |
| CC22(10) | t223 | 1 | V | Neg | |
| 1 | IVa | Neg | |||
| CC30(20) | t021 | 2 | IVa | Neg | |
| t318 | 1 | IVa | Neg | ||
| 1 | IVa | Pos | |||
| CC80(15) | t044 | 1 | IVa | Pos | |
| 2 | V | Pos | |||
| CC88(15) | t786 | 2 | IVa | Neg | |
| 1 | IVa | Neg | |||
| NA(5) | t11839 | 1 | V | Neg | |
| Saudi Arabian spa typed isolates | CC5(6.66) | t002 | 1 | V | Neg |
| CC8(13.33) | t008 | 2 | V | Neg | |
| CC15(26.66) | t084 | 1 | IVa | Neg | |
| 1 | V | Neg | |||
| t085 | 2 | V | Neg | ||
| CC22(6.66) | t223 | 1 | V | Neg | |
| CC25(13.33) | t349 | 1 | V | Neg | |
| t660 | 1 | IVa | Neg | ||
| CC80(6.66) | t376 | 1 | Nontypeable | Pos | |
| CC188(6.66) | t189 | 1 | V | Neg | |
| NA(6.66) | t442 | 1 | IVa | Neg | |
| NA(6.66) | t6836 | 1 | Nontypeable | Pos | |
| NA(6.66) | t11841 | 1 | IVa | Pos | |
MLST CC, multilocus sequence typing clonal complex (supposed allocations was based on the Ridom spa database and Monecke et al.23
This study, to the best of our knowledge, represents the first report on molecular characteristics of CA-MRSA recovered from anterior nares of outpatients attending health care centers in both Egypt and Saudi Arabia.13,14 We found a high carriage rate of S. aureus in EGOs and SAOs, whereas MRSA carriage rate was slightly higher in Egypt (32%) than in Saudi Arabia (25%). The high carriage rate is compatible with the reported worldwide trend in increasing CA-MRSA infections.11,12 In a recent report from Saudi Arabia, 26 studies on MRSA prevalence in five KSA regions were analyzed since 2002 to 2012. Consequently, the overall mean estimation of MRSA prevalence was 35.6%, where MRSA prevalence varied among regions. Surprisingly, there were no data regarding MRSA prevalence in Al-Qassim region.14,24 In Egypt, CA-MRSA carriage rate in our study was lower than the percentage of CA-MRSA (47.37%) in a sample of outpatients with skin and soft tissue infections who attended the dermatology department of a medical school in Alexandria university.25 In addition, other studies have reported MRSA prevalence in excess of 50%, but this study has mostly included hospital inpatients.25 On the other hand, the carriage rate observed in this study is higher than previously reported rates from other countries in the Middle East area.24,26
Several factors could have been responsible for the higher carriage rate of CA-MRSA including the overconsumption of antibiotics in both Egypt and Saudi Arabia, which is a contributing factor for multidrug resistant bacteria. Several reports have found a positive correlation between S. aureus resistance to methicillin and consumption of beta-lactam combinations.27,28 Other factors include overcrowding, poor hygiene, nutritional status, prevalence of parasitic infections, hot and humid weather, and presence of foreign labor and foreign tourists, which have different degrees of priorities in both countries.29
Among antimicrobial susceptibility phenotypes, a low percentage of isolates showed multiresistance. This was expected from community isolates. All Egyptian MRSA isolates were found to carry either type V or IVa SCCmec. The case was the same for Saudi Arabian MRSA isolates, but nearly 35% of the isolates remained nontypeable. This confirms that MRSA strains are community strains and confirm the tendency of CA-MRSA spreading among outpatients. We have found that PVL+ isolates were low in frequency, while PVL− isolates were predominant in both countries. This supports the hypothesis that PVL gene is not necessarily the key factor in determining CA-MRSA strains, and it does not play a greater role in increasing dissemination of these strains. Several studies have reported the predominance of PVL− strains among the CA-MRSA isolates.30–34
In this study, four clonal complexes were identified in both Egypt and Saudi Arabia including (I) CC5-V-PVL negative (t688) found in high rate (30%) in Egyptian MRSA isolates, while it was present with low incidence rate in Saudi MRSA isolates as (t002), CC5 was a common and widespread CA-MRSA clonal complex. It has been described in Australia, Ireland, Germany and Abu Dhabi (23), Israel with high incidence rate,35 Gaza Strip,36 and Tunisia.37 (II) CC8-V-PVL positive (t008) was found in Saudi Arabia with high incidence and as PVL positive and low incidence in Egypt. CC8 is a pandemic MRSA lineage, which is the predominant strain in the USA; CC8-MRSA-V has been reported in Pakistan,14 Europe,12 Denmark, Germany and Australia.23 (III) CC22-V-PVL negative, and CC22-IVa-PVL negative (t223), PVL-negative, CC22-IV represent a HA-MRSA pandemic strain known as UK-EMRSA-15, or Barnim Epidemic Strain. A recent report from Gaza Strip36 have highlighted the presence of CC22-MRSA-IV-PVL negative (t223) in 64% of MRSA isolates, and showed also the presence of CC22-V-PVL negative but in low incidence. This strain is common in Western Europe and has also been reported in many countries including Malta, Kuwait, Abu Dhabi, India,23 and Tunisia.37 Sporadic cases of a PVL-negative CC22-MRSA-V have been identified in Germany.23 IV.CC80-Iva-PVL+ (t044) has been found in Egypt at low incidence rate, CC80-PVL+ (t376) with nontypeable SCCmec has been found in Saudi Arabia also at low incidence rate. PVL-positive CC80-MRSA-IV has been described as the European CA-MRSA clone. This strain is widespread and has been isolated from different countries in Europe,12,23 Gaza strip,36 Kuwait14 and Tunisia.37
In Egypt, two more clonal complexes were observed, CC30-IVa-PVL-negative, PVL-positive (t021, t318) and CC88-IVa-PVL-negative (t786) with an incidence rate of 20% and 15%. CC30 is another important clonal complex from which CA-MRSA originate. A PVL-negative strain of ST30-MRSA-IV is sporadically isolated in Ireland and Australia.23 Another important CC30-MRSA strain is the PVL-positive ST30-MRSA-IV, Southwest Pacific Clone, which is widespread in Germany, Switzerland, the UK and Australia.23 Several different CA-MRSA belong to CC88, which appears, based on spa sequences and hybridization profile, closely related to CC1 and CC80. PVL-negative CC88-MRSA-VI has been identified sporadically in Western Australia,23 and has been recently reported in the Middle east; Gaza strip36 and Africa; Gabon, Gulf of Guinea.38,39
In Saudi Arabia, three more clonal complexes were observed: CC15-IVa-PVL-negative, CC15-V-PVL-negative (t084, t085) with a 27% incidence rate, CC15-MRSA has been detected in a collection of Italian MRSA strains isolated in 1980.23 It was reported in a recent study of S. aureus isolates from 16 of the most populous European countries with high incidence rates,12 Iran in Asia,40 and Gabon, Tunisia and Gulf of Guinea in Africa.37–39 CC25-IVA-PVL-negative, CC25-V-PVL-negative with a 13% incidence rate. CC25 was previously reported in Europe,12 Iran in Asia,40 and Gabon in Africa.39 ST188-MRSA-IV has been found sporadically in Australia, other ST188-MRSA have been observed in Asian countries: ST188-MRSA-III/spa t189 in Korea, PVL-negative CC188-MRSA-V in Hong Kong, and PVL-positive ST188-MRSA-V in Malaysia.23
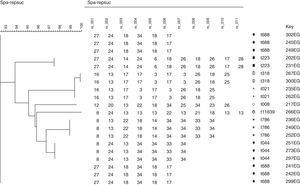
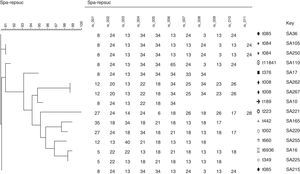
Spa typing reflects a high genetic diversity for MRSA isolates in Saudi Arabia comparing to that in Egypt. There was a close relationship observed when phylogenetic analysis of spa sequences executed for each country alone using the BioNumerics software version 7.1 (APPLIED MATHS, Austin, USA), surprisingly; Phylogenetic analyses of all spa types from both countries revealed a close relationship among both Egypt and Saudi Arabia MRSA isolates (Figs. 3–5). This may be referred to the high traveling rate from Saudi Arabia to Egypt and Vice versa which consider a way of transmission of different clones between countries.
In this study, we have tried to delineate molecular epidemiological features that MRSA isolates of outpatients have in common with those in other parts of the world, while highlighting unique features of both Egyptian and Saudi Arabian isolates. With global transmission of MRSA, the local epidemiology of both countries may be changing due to the introduction of new strains. Our study has some limitations concerning the extent of representation of the MRSA population in both Egypt and Saudi Arabia. First, our data were collected in health centers of specific locations in each country that might not be representative of isolates from different parts of each country. Second, despite the collection instructions, we were not able to categorize outpatients according to chronic or acute diseases they had, because such information was not provided by the physicians who collected nasal swabs.
In conclusion, MRSA still remains a significant public health problem. Surveillance is regarded as a means of generating ‘information for action’. In terms of MRSA, coordinated surveillance across Middle-east area is strictly needed, while information is still scarce in this area of the world. On the other hand, measures to control the spread of MRSA must be routinely applied. Furthermore, reducing the selective pressure for emergence and persistence of MRSA associated with overuse of antibiotics by improving antibiotic prescribing will improve the situation.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank Dr. Ghader Gad and her colleagues for their assistance in samples collection.