This study evaluated the use of polymerase chain reaction for cryptococcal meningitis diagnosis in clinical samples.
Materials and methodsThe sensitivity and specificity of the methodology were evaluated using eight Cryptococcus neoformans/C. gattii species complex reference strains and 165 cerebrospinal fluid samples from patients with neurological diseases divided into two groups: 96 patients with cryptococcal meningitis and AIDS; and 69 patients with other neurological opportunistic diseases (CRL/AIDS). Two primer sets were tested (CN4–CN5 and the multiplex CNa70S–CNa70A/CNb49S–CNb-49A that amplify a specific product for C. neoformans and another for C. gattii).
ResultsCN4–CN5 primer set was positive in all Cryptococcus standard strains and in 94.8% in DNA samples from cryptococcal meningitis and AIDS group. With the multiplex, no 448-bp product of C. gattii was observed in the clinical samples of either group. The 695bp products of C. neoformans were observed only in 64.6% of the cryptococcal meningitis and AIDS group. This primer set was negative for two standard strains. The specificity based on the negative samples from the CTL/AIDS group was 98.5% in both primer sets.
ConclusionsThese data suggest that the CN4/CN5 primer set was highly sensitive for the identification of C. neoformans/C. gattii species complex in cerebrospinal fluid samples from patients with clinical suspicion of cryptococcal meningitis.
Cryptococcal meningitis is one of the most serious systemic mycoses, mainly affecting immunocompromised patients and causing high morbidity and mortality. The course of infection is often a life-threatening disease that is typically observed in later stages of acquired immunodeficiency syndrome (AIDS).1–4
Cryptococcal meningitis affects approximately one million persons in the world each year and results in more than 400,000 deaths within three months after the onset of the disease.5 Latin America, as a global region, has the third highest incidence of the disease, with around 54,400 cases annually.6 Fortunately, in recent years, AIDS-associated cryptococcal meningitis has decreased dramatically in Brazil, which is a middle-income country with universal access to antiretroviral therapy.4,7 However, until today, cryptococcal meningitis remains a common complication in Brazil and other Latin American countries as it represents the primary cause of opportunistic meningitis and the second-most frequent neurologic opportunistic infection in HIV-infected patients.6,7
Patients with cryptococcal meningitis usually present severe neurological manifestations, particularly secondary to intracranial hypertension.7–9 A rapid laboratorial diagnosis is extremely important to identify Cryptococcus species.10 Routinely, microscopic examination, cryptococcal polysaccharide antigen, and culture of cerebrospinal fluid (CSF) samples lead to the diagnosis.
Several studies have demonstrated the use of molecular methods for determining the genetic characteristics of Cryptococcus species.11–15 Nevertheless, there is scarce information about the use of molecular diagnosis in clinical samples. Molecular methods such as polymerase chain reaction (PCR) potentially identify the fungus and the genotype of the strain.
Considering that no method is entirely effective in determining the presence of this pathogen, it is helpful to diagnose on the basis of results obtained from different methodologies to rule out false-negative or false-positive results. Hence, molecular methods such as PCR have proven to be useful for this purpose, since they can provide a definitive diagnosis as well as evaluation of specific treatment.
The aim of this study is to evaluate the use of PCR for molecular diagnosis of cryptococcal meningitis.
Materials and methodsThe sensitivity and specificity of the methodology were evaluated using Cryptococcus reference strains in the first part and CSF samples from patients with neurological diseases in the second part of the study.
Patients and clinical samplesThe assays were evaluated analyzing 165 CSF samples with commonly used, well-known laboratory and clinical diagnoses. The clinical samples were divided into two groups. Group cryptococcal meningitis and AIDS (CM/AIDS) consisted of 96 CSF samples from patients with CM/AIDS. Cases of cryptococcal meningitis were confirmed using positive microscopic examination by India ink and latex agglutination of CSF samples.16 All patients were admitted to the Instituto de Infectologia Emilio Ribas, Sao Paulo, SP, Brazil. The CRL/AIDS group (control) consisted of 69 CSF samples from patients with confirmed diagnosis (by clinical, radiological and laboratory features) of other neurological opportunistic infections including cerebral toxoplasmosis, progressive multifocal leukoencephalopathy, HIV-associated neurocognitive disorders, tuberculous meningitis, and neurosyphilis.
Ethical considerationsAll patients provided written informed consent. The institutional review board of both Instituto Adolfo Lutz and Instituto de Infectologia Emilio Ribas committees approved this study.
Laboratory diagnosisAt the Instituto de Infectologia Emilio Ribas the CSF samples were collected in aseptic conditions, immediately transferred to sterile tubes, sent to laboratory and processed. Each sample was split in two tubes. One of them was used for microscopic examination by India ink and latex agglutination. The other was sent to the Instituto Adolfo Lutz for molecular diagnosis. Microscopic examinations by India ink was conducted as previously described.17,18 CSF samples were centrifuged at 3000×g for 10min. One drop of the pellet was used for microscopic observation after India ink preparation. The polysaccharide capsule antigen was detected in CSF samples using a latex agglutination Kit (Cryptococcus Antigen Latex Agglutination Test–Immy) according to the manufacturer's instructions.
Yeast preparationThe assays were also determined using DNA extracted from standard strains representing each molecular type including: WM 148 (serotype A, VNI), WM 626 (serotype A, VNII), WM 628 (serotype AD, VNIII), WM 629 (serotype D, VNIV), WM 179 (serotype B, VGI), WM 178 (serotype B, VGII), WM 175 (serotype B, VGIII), and WM 779 (serotype C, VGIV).12 The strains were prepared as previously described.12 Briefly, each isolate was plated on Sabouraud dextrose agar and incubated at 30°C for 48h. The yeast cells were transferred to a microcentrifuge tube containing 1mL of 50mM EDTA (Sigma Chemical), pH 8.0, mixed, and centrifuged at 13,000×g for 15min. The supernatant was removed, and the pellet was resuspended in 200μL of 50mM EDTA, pH 8.0, containing 40μL of lysing enzyme from Trichoderma harzianum (10mg/mL) (Sigma Chemical), and then incubated at 37°C for 2h. Subsequently, this mixture was centrifuged at 13,000×g for 10min, and the pellet was dissolved in lysis buffer containing 10mM Tris–HCl, pH 8.0; 10mM EDTA, pH 8.0; 0.5% sodium dodecyl sulfate; 0.01% N-laurilsarcosyl; and 100mg/mL proteinase K. The mixture was vortex mixed and incubated at 56°C for 2h.
DNA purification and PCRDNA molecules of protoplasts or CSF samples were extracted by QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions. CSF samples were previously centrifuged for 15min at 9000×g. Packed cells were washed twice in PBS to prevent the action of any Taq polymerase inhibitor. Whole cells were incubated for 15min at 100°C in 200μL of ultrapure water containing four to five 1.5mm glass beads. Next, the cells were processed in a TissueLyser disruptor (Qiagen) for two cycles of 3min. After DNA extraction, the purity was determined by the ratio of O.D. at 260 and 280nm in a NanoDrop ND100 (Thermo Fisher Scientific, Waltham, MA, US).
The amplifications were carried out with a kit purchased from Promega (Go Taq Green Master Mix). The PCR mix (12.5μL) consisted of 1 unit of Green GoTaq® DNA polymerase, 10mM Tris–HCl, pH 8.5; 50mM KCl; 1.5mM MgCl2; and 200mM of each dNTP. The reactions included the PCR mix, 10μL of each DNA template and 25pmol of each primer to a final volume of 25μL. The amplifications were performed in an automated thermal cycler and each run contained one negative control (ultra-pure water) and one positive (DNA extracted from serotype A, VNI culture). After thermal cycles, PCR products were electrophoresed in 2% agarose gels in TBE buffer and stained with ethidium bromide. The size of fragment was based on a comparison with a 100-bp ladder.
Primer set selectionThe experiments were carried out by using two primer sets. The first set was the CN4–CN5 (5′ATCACCTTCCCACTAACACATT3′ and 5′GAAGGGCATGCCTGTTTGAGAG3′), which amplified a 136-bp sequence from a specific gene coding region for rDNA of C. neoformans as target.15 The amplifications were performed by one initial denaturation cycle for 5min at 94°C, 30 cycles of denaturation at 94°C for 1min, annealing at 55°C for 1min, and extension at 72°C for 1min. The procedure was completed by a final cycle extension for 10min. The second was a multiplex PCR. The CNa70S–CNa70A (5′ATTGCGTCCACCAAGGAGCTC3′ and 5′ATTGCGTCCATGTTACGTGGC3′) amplified a 695-bp fragment of a region on C. neoformans chromosome 3 that includes the coding sequence of a putative aminotransferase gene. The CNb49S–CNb-49A (5′ATTGCGTCCATCCAAGGTGTTGTTG3′ and 5′ATTGCGTCCATCCAACCGTTATC3′) amplified a 448-bp fragment of a region on C. gattii chromosome 2, which includes the coding sequence of a putative polymerase.14,19 The PCR products were amplified by one initial denaturation cycle for 5min at 94°C, 35 cycles of denaturation at 94°C for 45s, annealing at 65°C for 60s, and extension at 72°C for 60s. The procedure was completed by a final cycle extension for 10min. To control the course of extraction and check for PCR inhibitors, all samples were assayed using the primer set β1–β2 (5′ACCACCAACTTCATCCACGTTCACC3′ and 5′CTTCTGACACAACTGTGTTCACTAGC3′),20 which amplified a 140-bp fragment of the human β-globulin gene. The reactions using the human primer set were run simultaneously with the same temperature protocol for the CN4–CN5 primer set and in the same PCR machine.
Data analysis and quality assuranceSeparate rooms were used for: (1) DNA extraction; (2) PCR mix and primer preparation; (3) adding DNA from clinical samples and positive control; and (4) post-PCR agarose gel electrophoresis analysis. DNA clinical samples were assayed in duplicate and, at least, twice. Clinical and laboratory (microscopic examination by India ink and latex agglutination) diagnoses were the gold-standard to establish sensitivity, specificity, positive (PPV) and negative (NPV) predictive values, which were calculated as: (i) sensitivity – ratio of true positives/(true positives+false negatives)×100; (ii) specificity – ratio of true negatives/(true negatives+false positives)×100; (iii) PPV – ratio of true positives/(true positives+false positives); and (iv) NPV – ratio of true negatives/(true negatives+false negatives).
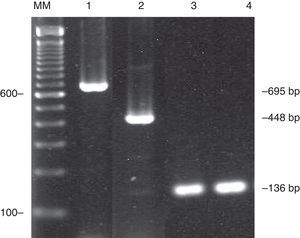
ResultsTwo PCR primer sets to determine C. neoformans/C. gattii species complex in clinical samples were evaluated since they amplified different genetic regions. Initially the primer sets were evaluated for sensitivity using the standard strains. CN4–CN5 primer set amplified the 136-bp product in all standard strains tested. The same DNA samples were also tested for CNa70S–CNa70A/CNb49S–CNb-49A multiplex. Four standard strains (serotypes A-VNI and VNII, serotype AD-VNIII, serotype D-VNIV) amplified the 695-bp fragment; and two (serotype B-VGII and serotype C-VGIV) amplified the 448-bp fragment. No amplified product was shown in two standard strains (serotypes B-VGI and VGIII) (Table 1). These experiments were conducted three times and, as an illustration, Fig. 1 shows the amplified PCR products of both primer sets used in this study. Both did not amplify other yeasts such as Candida albicans, C. parapsilosis, C. tropicalis, and C. glabrata.
Evaluation of primer sets CN4–CN5 and CNa70S–CNa70A/CNb49S–CNb-49A in PCR for Cryptococcus standard strains.
| Standard strains | Primer sets | |
|---|---|---|
| CN4–CN5 | CNa70S–CNa70A/CNb49S–CNb-49A | |
| WM 148 (serotype A, VNI) | Positive | Positive |
| WM 626 (serotype A, VNII) | Positive | Positive |
| WM 628 (serotype AD VNIII) | Positive | Positive |
| WM 629 (serotype D, VNIV) | Positive | Positive |
| WM 179 (serotype B, VGI) | Positive | Negative |
| WM 178 (serotype B, VGII) | Positive | Positive |
| WM 175 (serotype B, VGIII) | Positive | Negative |
| WM 779 (serotype C, VGIV) | Positive | Positive |
Amplified PCR products of two different target regions of Cryptococcus neoformans/C. gattii species complex. CNa70S–CNa70A/CNb49S–CNb-49A multiplex amplified a 695-bp product of C. neoformans (line 1) and 448-bp of C. gattii (line 2). CN4–CN5 primer set, amplified a 136-bp product (lines 3 and 4). The DNA fragments were resolved in 2% agarose gels stained with ethidium bromide. Lane MM, 100-bp ladder.
The good quality of all 165 DNA samples was confirmed by the amplification of β1–β2 primer set that amplified a PCR fragment of human β-globulin gene of 140-bp. Positive amplifications showed that no substance present in DNA samples inhibited the reactions.
Next, the sensitivity and specificity of the CN4/CN5 and the CNa70S–CNa70A/CNb49S–CNb-49A multiplex were determined in DNA samples extracted from CSF samples of both groups of patients. Table 2 shows the results of each group in detail. The analysis of the clinical samples from the CM/AIDS group revealed that 94.8% of these samples were positive (91 out of 96) using the CN4/CN5 primer set. The specificity of the assay, based on the analysis of all 69 negative samples from CTL/AIDS group was 98.5%.
Summary of the results using CN4–CN5 and CNa70S–CNa70A/CNb49S–CNb-49A primer sets in PCR using CSF samples from AIDS patients.
| Primer sets | PPVa | NPVb | PCR resultsNumber of samples (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Cryptococcal meningitis | Other neurological diseases | |||||||
| Positivec | Negative | Total | Positive | Negatived | Total | |||
| CN4–CN5 | 0.99 | 0.93 | 91 (94.8%) | 5 | 96 | 1 | 68 (98.5%) | 69 |
| CNa70S–CNa70A | 0.99 | 0.53 | 62 (64.6%) | 34 | 96 | 1 | 68 (98.5%) | 69 |
According to the multiplex CNa70S–CNa70A/CNb49S–CNb-49A, no 448-bp product of C. gattii (CNb49S-CNb-49A) was shown in the clinical samples of either group. The 695-bp products of C. neoformans were shown in only 64.6% (62 out of 96) of the CM/AIDS group. In CTL/AIDS, the specificity was also 98.5%. Only one sample out of 69 had a false-positive result. Although the specificity was good, the sensitivity was very low as well as the NPV (0.53).
DiscussionDespite the observed decline of cryptococcal meningitis in AIDS patients, its occurrence still represents a determinant of poor prognosis in HIV-infected patients.6 Additionally, this fungal infection in the central nervous system leads to severe neurological complications. In this way, its successful treatment depends on rapid and accurate identification of the causative agent.16 Consequently, accurate and rapid diagnosis is critical. In recent years, great advances have been made in developing laboratory techniques to diagnose infectious diseases. Nevertheless, despite the well-established diagnosis of cryptococcal meningitis some aspects of the diagnosis remain challenging and there are still gaps that need to be addressed. For instance, a negative result from an in vitro culture might be due to the presence of nonviable or fastidious yeast in the sample. Thus, the use of PCR in CSF samples associated with methods already used at the laboratory could increase the diagnostic efficacy.
Several studies based on molecular methods have shown that C. neoformans isolates exhibit different degrees of genetic heterogeneity among clinical and environmental isolates. The primer sets used in these Cryptococcus studies were normally used for genotyping characterization.11,21–25 However, there have been few studies evaluating the efficacy of such methodology in molecular diagnosis. Here, we chose two previously described primers’ set widely used for Cryptococcus genotyping. Two primer sets were tested as they amplify different genetic regions. The CN4–CN5 primer set amplified a 136-bp sequence from a specific region of gene coding for rDNA as target.15
The sensitivity, accuracy and specificity of a diagnostic test depend also on optimal working conditions. In order to improve the PCR sensitivity, the CSF samples should be processed rapidly within 48h of collection to prevent Taq polymerase inhibition that could modify PCR results.26 In addition, the DNA extraction and PCR inhibitors may be evaluated by a primer set that amplified human PCR fragments. In this study, positive amplifications showed that no substance present in DNA samples inhibited the reactions.
Firstly, the efficacy of both primer sets was analyzed using Cryptococcus standard strains and then the clinical samples. The CN4–CN5 primer set was highly sensitive since it amplified all Cryptococcus standard strains used in this study. These data were further confirmed using 96 clinical samples from the CM/AIDS group that revealed good sensitivity (94.8%) and negative predictive value of 0.93. Five cases of the CM/AIDS group were false negatives. The PCR results could be due to low or non-uniform distribution of specimens from which the DNA was extracted. Furthermore, the specificity was also good (98.5%) with high positive predictive value (0.99). Only one DNA sample from CLR/AIDS had a false-positive result. Similar results have been reported previously.27
The use in molecular diagnosis of a multiplex primer set could be an excellent alternative, since in the same reaction it could be possible to detect the organism and, at the same time, genotype the infecting strain. For this purpose, the CNa70S–CNa70A/CNb49S–CNb-49A multiplex was tested in this study. According to previous studies, 695-bp PCR products were shown in C. neoformans strains and a 448-bp fragment of C. gattii.14,19 In this study, the PCR using the (CNa70S–CNa70A/CNb49S–CNb-49A) multiplex was unable to amplify two Cryptococcus standard strains (serotypes B-VGI and VGIII). In addition, no 448-bp product of C. gattii was shown in the clinical samples of either group. These data could suggest that the majority of the DNA samples from CM/AIDS had yeast belonging to C. neoformans. However, the 695-bp products of C. neoformans were shown only in 64.6% of the CM/AIDS group resulting in low sensitivity with very low NPV (0.53). In the CM/AIDS group, 34 DNA samples had false-negative results.
Collectively these data suggest that although the multiplex primer set could detect the organism and, at the same time, genotype the infecting strain, the sensitivity was too low. On the other hand, the primer set CN4/CN5 was highly sensitive for C. neoformans/C. gattii species complex determination in CSF samples from patients with clinical suspicion of cryptococcal meningitis or the treatment control.
Conflicts of interestThe authors declare no conflicts of interest
This study was supported by grants from: (i) FAPESP (Fundaçao de Amparo à Pesquisa do Estado de São Paulo, Brazil), Proc: 2011/13939-8, (ii) CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), Proc: 303489/2012-0. Jim Hesson of AcademicEnglishSolutions.com revised the English.