One approach to identifying HIV-1 vaccine candidates is to dissect the natural antiviral immune response in treatment-naïve individuals infected for over ten years, considered slow progressor patients (SPs). It is suspected that SP plasma has strongly neutralizing antibodies (NAb) targeting specific HIV viral epitopes.
MethodsNAbs levels of 11 HIV-1-infected SPs were detected by PBMC-based neutralization assays. To investigate SP NAb epitope, this study used a biopanning approach to obtain mimotopes of HIV-1 that were recognized by SP plasma NAbs. IgG was purified from high-titer NAb SP plasma, and used as the ligand for three rounds of biopanning to select HIV-specific mimotopes from a phage-displayed random peptide library. Double-antibody sandwich ELISA, competitive inhibition assays, and peptide sequence analysis were used to evaluate the characteristics of phage-borne mimotopes.
ResultsSPs had significantly more plasma neutralizing activity than typical progressors (TPs) (p=0.04). P2 and P9 plasma, which have highest-titer HIV-NAb, were selected as ligands for biopanning. After three rounds of biopanning, 48 phage clones were obtained, of which 22 clones were consistent with requirement, binding with HIV-1 positive plasma and unbinding with HIV-1 negative plasma. Compared with linear HIV-1 protein sequence and HIV-1 protein structure files, only 12 clones were possible linear mimotopes of NAbs. In addition, the C40 clone located in gp41 CHR was found to be a neutralizing epitope, which could inhibit pooled HIV-1 positive plasma reaction.
ConclusionBiopanning of serum IgG can yield mimotopes of HIV-1-related antigen epitopes. This methodology provides a basis for exploration into HIV-1-related antigen-antibody interactions and furthers NAb immunotherapy and vaccine design.
A collaborative global effort is ongoing to develop an effective human immunodeficiency virus (HIV) vaccine.1–4 However, HIV-1 vaccine design has faced numerous difficulties. The main problem is the ability of HIV to rapidly generate mutants resistant to immune responses. Considering the complexity of HIV-1 replication and pathogenesis, it is widely acknowledged that an HIV-1 vaccine, in order to be effective, should include multiple antigens and should be able to generate strong and broad neutralizing antibodies (NAbs), as well as cell-mediated immune responses.
NAbs are considered a key component of protective vaccines against HIV-1; however, despite tremendous efforts, vaccination attempts have thus far failed to induce broad neutralization activity. At present, only a small number of broadly monoclonal NAbs have been isolated from HIV-positive individuals.5 These are the few NAbs that have exhibited potent and broad HIV-1-neutralizing activity in vitro and have been shown to prevent HIV-1 infection in animal models.6–8 Of these isolated antibodies, four antibodies in particular, IgG1b12, 2G12, 2F5, and 4E10, have been proven to be the most effective.9–13 Recently, these NAbs have been administered in acutely and chronically HIV-1-infected patients in order to assess their potential to delay viral rebound after structured treatment interruption.14,15
The ability to discover immunogens that are able to elicit NAbs can be facilitated by the exploration of the interactions of these antibodies with the HIV-1 envelope glycoprotein (Env).6 Meanwhile, increased understanding of NAb epitopes may help to design immunogens capable of inducing such antibodies upon immunization. The identification of new broadly cross-reactive HIV-1 NAbs and their conserved epitopes is therefore of obvious importance for the development of HIV drugs and vaccines. Many antibodies directly target the HIV-1 Env, because they are the only virus-encoded determinants that are present on the virus surface. Env is comprised of two glycoproteins, gp120 and gp41, which mediate cell fusion in the first step of HIV-1 infection. Antibodies targeting conserved regions of gp120 and gp41 may be able to neutralize a wide array of circulating HIV-1 isolates and therefore constitute potential targets for therapeutic intervention.16 A vaccine could represent a strategy to induce high levels of NAbs with predefined epitope-specificity against HIV-1.
Natural infection with HIV-1 generates many virus-specific antibodies, but most are non-NAbs.17 Interestingly, a group of HIV-infected patients has been able to control the infection without antiviral therapy, and have been termed slow progressors (SPs). SP patients do not show any evidence of immunological deterioration or clinical symptoms of immunodeficiency despite being infected for over ten years without any antiretroviral therapy. Multiple factors have been reported to contribute to the SP status, including deletions in viral regulatory genes or the CCR5 coreceptor gene, as well as several immunological factors. Studies have reported that SP plasma facilitates neutralization of primary HIV-1 isolates more effectively than plasma from HIV-1 typical progressors (TPs).18 Moreover, previous studies by the authors have shown that the plasma NAb titer of SPs is higher than that of TPs (unpublished data). If NAbs in SPs are indeed involved in delaying disease progression, a greater comprehension of their epitopes might assist in designing immunogens. SP sera are also rich in virus-specific antibodies, which is advantageous for the process of biopanning.
Biopanning is a powerful and versatile approach that allows for specific desirable epitope-antibody interaction.19,20 Biopanning commonly requires the iterative application of phage display technology, which due to its screening capacity and broad applicability occupies a central role in the study of protein-protein interactions.21–23 The power of phage display lies in the direct physical linkage between a phenotype and its genotype. A library of bacteriophages, each encoding a random short peptide that is displayed on the viral surface, is allowed to interact with a defined target protein. Through targeted binding and elution, peptides capable of interacting with the target can be easily selected from a milieu of millions and identified by sequencing the DNA encapsulated in the corresponding phage particle.24–26 Biopanning combines current protein and peptide expression and modification methods with simple microbiological procedures that enable fast amplification of hundreds of selected peptides, which can be applied to retrieve collections of relevant epitopes and mimotopes binding to antibodies of interest.
In this study, phage displayed peptide libraries were screened with IgGs from SP plasma. The objective was to identify the epitopes of NAbs, as these should represent immunogens potentially able to indicate antigens that are capable of inducing antibodies upon immunization for HIV-1 vaccine.
Materials and methodsStudy populationThirty-four HIV-1-infected individuals from Henan province were enrolled in this study. This study was conducted in accordance with the Declaration of Helsinki, with the approval of the Ethics Committee of China Medical University. An informed consent was obtained from all participants. All patients acquired HIV-1 infection via contaminated blood transmission. HIV-1 infection was screened by anti-HIV ELISA (Vironostika, Organon Tenika – The Netherlands) and confirmed by Western blot (Genelab Diagnostics – Singapore). Among these patients, 11 were known SPs (with asymptomatic and stable CD4+ T-cell counts ≥500 cells/μL for at least ten years), while 23 were TPs (CD4+ T cell counts <500 cells/μL for at least five years). Only antiretroviral (ARV) treatment-naïve patients were included in this study. HIV-1 subtype was determined by phylogenetic analysis based on sections of env, pol, and gag regions. All SP and TP patients were infected with HIV-1 B’-clade. Plasma from HIV-negative individuals samples were used as a negative control in biopanning and phage ELISA assays.
Neutralization assayPBMC-based assays using SF33 strains were performed in 96-well culture plates by incubating virus stock with serial diluted heat-inactived plasma specimen. Approximately 500 TCID50 in 50μL (determined by Spearman-Karber equation assay) of SF33 were incubated with 50μL diluted plasma for 30minutes at 37°C in a well of a 96-well plate, followed by the addition of PHA-activated PBMC (1.0×105 in 100μL of complete RPMI 1640 medium). After an overnight incubation at 37°C, 150μL of the culture supernatant were removed, followed by the addition of 150μL of fresh culture medium. On the fourth day. 100μL of the culture supernatant was exchanged with 100μL of fresh culture medium. Seven days after infection initiation, duplicate samples were harvested for measurement of p24-Ag by ELISA. IC50 values for patient serum NAb activity were calculated, and the SP and TP IC50 values were compared by Student's t-test.
The PBMCs used for this assay were isolated from healthy donors by standard density gradient centrifugation with Histopaque-1077 (Sigma). Isolated cells were plated in 25-cm2 plastic flasks and incubated at 37°C for two hours. Non-adherent cells were collected and resuspended at 1×106 cells, and were stimulated in 10mL RPMI-1640 medium containing 10% FBS, 5μg/mL PHA, and 100 U/mL IL-2 (Sigma) by incubation at 37°C for three days.
Biopanning of phage displayed peptide libraryThe biopanning of phage-displayed peptide was performed as described elsewhere.27 Briefly, Dynabeads M-280 Tosylactivated were resuspended by pipetting. Goat anti-human IgG antibody was dissolved in 0.1M H3BO3 (pH 9.5). Dynabeads were precoated with goat anti-human Fc-specific antibody, according to the manufacturer's instructions, and were incubated with patient plasma for two hours at room temperature. Beads were washed six times with PBS/0.1% Tween-20 and continually rotated overnight at 4°C with 10μL (1.5×1011–2×1011 particles) from the original M13 bacteriophage peptide libraries displaying linear peptide chains consisting of 12 amino acids (Ph.D.TM – Phage Display Peptide Library; New England Biolabs). The next day, the beads were washed extensively with PBS/0.1% Tween-20, and bound phages were eluted by pH shift [0.2 Mglycine-HCl (pH 2.2)/1mg/mL BSA], neutralized with 1M Tris-HCl (pH 9.1). Beads were used for three rounds of selection. Positive selection was first performed with plasma from HIV-infected patients. Supernatants of the positive selection were amplified in Escherichia coli ER2783, precipitated overnight at 4°C, and used for two more rounds of selection. Negative selection was performed as the third step with pooled plasma from HIV-negative subjects. The plasma of patients was obtained respectively from SP patients P2 and P9. Tween-20 concentrations were increased stepwise (0.1, 0.3, 0.5%) during the three rounds of panning. After the third selection, the phages were titered and picked randomly. Plaques were picked from plates with no more than 100 clones for incubating and sequencing. Amplified phages were concentrated by PEG/NaCl precipitation and analyzed for specificity by ELISA. Single-stranded DNA of positive phage clones (OD values >2.0 control) were prepared and sequenced to deduce the peptide insert. DNA sequences encoding the peptides (170bp) were amplified with the primers PHD1-(5′-TTC GCA ATT CCT TTA GTG-3′) and PHD2- (5′-TTT GTC GTC TTT CCA GAC GT-3′). The PHD2 primer is recommended for dideoxy sequencing. Phage clone peptide sequences were determined and analyzed for linear homology to HIV-1 proteins by local BLAST. Phage peptide sequences were compared against a reference HIV-1 subtype B Env protein sequence (B.TH.90.BK132 ACC AY173951), using the Bioedit program to identify neutralization epitopes.
Detection of HIV-specific phage clonesPhages displaying mimotopes were subsequently tested with phage ELISA to confirm HIV specificity. Nunc-Immuno Maxisorp 96-well plates (Nalge-Nunc International) were coated overnight at 4°C with 100μL/well of M13 antibody (Pharmacia) at a concentration of 10μg/mL. The next day, the plates were blocked with 200μL blocking solution of 3% BSA in PBS for two hours at 37°C, and were washed three times in an automated plate washer. 100μL/well of phages, selected through biopanning, were added in a 1:1 ratio to wells with 100μL/well of a 1:100 dilution of plasma of either pooled HIV-1 negative controls or pooled HIV-1 positive controls. Mixtures were performed in triplicate and were incubated overnight at 4°C. The plates were washed, incubated with 100μL/well HRP-goat anti-human IgG conjugate for one hour at 37°C developed with ABTS substrate (Pharmacia) for 15minutes at room temperature, and read at 405nm. Clones that reacted with HIV-positive serum (OD value >2.0 fold blank control) were selected for further experimentation, while clones that reacted with HIV-negative serum (OD value >1.5 fold blank control) were discarded.
Competition inhibition binding of phage cloneTo determine whether phage clones represented mimotopes of gp41, competitive inhibition assays using gp41 were conducted. Nunc-Immuno Maxisorp 96-well plates were coated with 100μL/well gp41 peptide at concentration of 0.1μg/mL, overnight at 4°C. The next day, plates were blocked with 200μL blocking solution of 3% BSA in PBS for one hour at 4°C, and were washed three times with PBST (1% Tween-20). Plates were incubated with phage clone, which was diluted four-fold serially from 1:4 to 1:64, and pooled HIV-positive plasma (1:800) for two hours at 37°C. Samples were run in duplicate. After four washes with PBST, 100μL of a 1:5000 dilution of HRP-conjugated goat anti-human IgG was added and incubated for one hour at 37°C. After four washes with wash buffer, assays were developed at 37°C for 15 to 30minutes with ready-to-use TMB substrate, and read at 450nm.
Amplification and sequencing of gp41 fragmentHIV-1 RNA from plasma was extracted with QIAamp Viral DNA and RNA Mini Kit (Qiagen – Germany), according to the manufacturer's recommendations. RNA was reverse transcribed into cDNA with a reverse transcriptase kit (TAKARA – Japan). Two separate plasmas were obtained from patients P2 and P9. The HIV-1 gp41 gene (461bp) was amplified with outer primers gp41-1 (5′-TCT TRG GAG CAG CAG GAA GCA CTA TGG G- 3′HIV-1HXB2 7789-7816nt) and gp41-2 (5′-AAC GAY AAJ GGT GAR TAT CCC TGC CTAA- 3′HIV-1HXB2 8374-8347nt), and followed by nested PCR with inner primers gp41-3 (5′-ACA ATT ATT GTC TGG TAT AGT GCA RCA GCA - 3′HIV-1HXB2 7850-7879nt) and gp41-4 (5′-TTA AAC CTA YCA AGC CTC CTA CTA TCA TTA - 3′HIV-1HXB2 8310-8281nt). Purified PCR products were directly sequenced by dideoxy chain termination with primers gp41-5 (5′-CAA TTA TTG TCT GGT ATA GTG C-3′, HIV-1HXB2 7851-7872nt) and gp41-6 (5′-CAA GCC TCC TAC TAT CAT TA-3′ HIV-1HXB2, 8300-8281nt), on an automated sequencer 377 (Applied Biosystems - USA). DNAStar was used to predict conformational epitopes of the gp41 region. The two HIV-1 gp41 sequences used in this analysis containing neutralizing epitope have been submitted to Genbank under the following accession IDs: EU569806, EU569820.
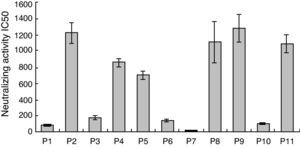
ResultsNeutralizing activity of SP and TP plasmaTo confirm NAbs as a likely factor in viral control in SP patients, in vitro neutralization assays with SP and TP plasma were performed and compared. Neutralization assay IC50 values for SP and TP groups are shown in Table 1. The neutralizing ability of SP plasma was significantly higher than that of TP patient plasma. SP patient mean IC50s are summarized in Fig. 1. All SP plasma samples neutralized the heterologous HIV-1 subtype B strain SF33. Plasma from SP patients P2 and P9 displayed strongly neutralizing activity, confirming that NAbs indeed contributed to strong neutralization in these patients.
Relevant HIV-1-specific epitopes were investigated through selection of mimotopes by biopanning. Biopanning with phage libraries screened with immobilized IgG from plasma from P2 and P9 yielded 48 phage clones, which were analyzed for specificity through phage ELISA. Clone selection was based on reactivity; 38 clones that reacted with the HIV-positive plasma pool were selected, subsequently, 16 clones of 38 clones that reacted with the HIV-negative plasma pool were discarded.
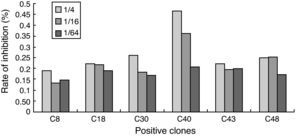
Analysis of HIV-1 mimotopesThe 22 phage clone peptide sequences were compared with linear HIV-1 protein sequence and HIV-1 protein structure files. 12 clones were determined to be possible continuous linear mimotopes of NAbs (Table 2). The remaining ten epitopes did not align with the reference sequence and may possibly represent discontinuous epitopes. Of the 12 linear clones, six clones mimicked epitopes on the envelope glycoprotein gp120. Clones 14, 15, and 31 were identical and were located in the gp120 C1 region. Clone 10 was located in the gp120 V1 loop, clone 22 occurred in the gp120 C2 region, and clone 23 was located in the gp120 V4 loop. Another six clones mimicked epitopes on gp41. Notably, clone 40 had the highest titer by inhibition ratio against gp41 of all six gp41 clones analyzed (Fig. 2). The sequence of clone 40 was localized in the gp41 C-terminal heptad repeat.
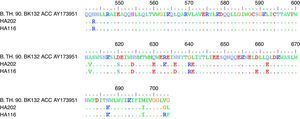
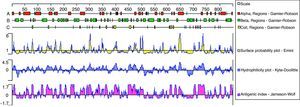
In order to identify conserved mimotopes that are capable of reacting with more than one HIV-positive plasma sample, phage clones were analyzed for cross-reactivity with a panel of plasma from HIV progressors by ELISA. The broadest cross-reactivity was observed in the phage clones representing immunodominant linear mimotopes, such as the gp41 CHR domain. Due to the fact that the C40 phage clone had a high inhibition ratio (46.7%) when reacted with HIV-positive plasma pools in competition ELISA, the P2 and P9 gp41 fragments were sequenced (Fig. 3). C40 corresponded to a region of the gp41 reference sequence residues 637-648. Compared with other HIV subtype sequences, the region that C40 mimicked was found to be relatively conserved (Table 3). In silico predictions of the gp41 region mimicked by C40 showed that the region was characterized by hydrophobicity and high antigenic index (Fig. 4).
The discovery of novel broadly cross-reactive human monoclonal NAbs to various antigens has major implications for the development of vaccines. Accumulating evidence has clarified the importance of broadly NAbs in enhancing the control of viral replication through cellular immune response. Previous reports of the presence of NAbs in SPs concluded that the SP response is broader in magnitude and breadth than the TP response.18 However, in most of these studies, SP patients were compared to patients with advanced AIDS. Therefore, the relatively superior neutralization of the SP plasma that was observed could be due to the lack of antibodies typically seen as a symptom of AIDS.
In this study, an experimental strategy that could facilitate the selection of high-titer antibodies from SPs was described. Screening of HIV-infected patient plasma with phage-displayed random libraries can identify peptide mimics of HIV-1 immunodominant epitopes that can extend the range of structural or protein-protein interaction studies of viral epitopes. In this study, phage display and biopanning were used in a reverse vaccinology approach to identify antigenic epitopes relevant for HIV-1 vaccine development. Eleven known SPs who had successfully controlled HIV-1 infection for more than 10 years were selected for research. Two of the 11 SP patients with the highest plasma NAb IC50 values were chosen for selection and characterization of mimotopes. Phage-displayed peptide libraries with immobilized IgG from two SPs were described, and 22 mimotopes were selected to be analyzed for homology to the HIV-1 Env protein. SP plasma was used to screen phage libraries for HIV specificity, resulting in the selection of 12 linear mimotopes, which mostly corresponded to immunodominant epitopes in the HIV-1 Env complex proteins gp120 or gp41. The overrepresentation of Env in isolated mimotopes can be explained by the fact that the Envs gp120 and gp41 are the only viral proteins exposed on the exterior of the virus particle; thus, they are the only proteins that elicit HIV-specific NAbs in infected individuals. Vaccine design efforts have thus focused on eliciting broad NAb response to these two proteins.
The methods used in this study only surveyed linear epitopes; discontinuous conformational epitopes for broadly neutralizing activity may have been generated, but were not confirmed. Conserved linear domains on gp41 have been observed, and for this reason gp41 was chosen to survey epitopes for novel targets for Nabs.28,29 Competition ELISA showed phage clones that represented epitopes on gp41, including the high-titer clone C40, which had a high inhibition ratio (46.7%) against a pool of HIV-positive plasma. A consensus motif C40 that mimics the gp41 CHR epitope was also identified. The HIV-1 transmembrane subunit gp41 is an important modulator in HIV-1 entry in its host cell that contains a hydrophobic, glycine-rich sequence, referred to as the fusion peptide at its amino terminus. The fusion peptide region is followed by two heptad repeat sequences of hydrophobic amino acids (N-terminal heptad repeat and C terminal heptad repeat) that form helical domains with hydrophobic faces. Packing interactions between these two regions are critical for membrane fusion.16,20,30–33 In the prehairpin intermediate state of gp41, the N-terminal heptad repeat (N-HR, residues 542–591 of HIV-1 Env) and the C-terminal heptad repeat (C-HR, residues 628–661) interact with another exposed N-HR to form a trimeric coiled-coil.30,34–36 The C40 clone mimicks an epitope on the internal trimeric coiled-coil of the C-HR.
The present results also suggest that the functionally important gp41 structure is not masked as efficiently in SP viruses, resulting in better recognition and generation of anti-gp41 antibodies with broader neutralizing capacity. For the conformational epitopes predicted by DNAStar, it was also observed that the region of gp41 mimicked by C40 was characterized with hydrophobicity and a high antigenic index. Hence, C40 may be used for further investigation as an engineered immunogenic version of its gp41 epitope. In addition, C40 was located in the fusion-inhibitor drug enfuvirtide (T20) target region in the gp41 heptad repeat 1 (HR1) domain.37 T20 is a member of a successful new generation of entry inhibitors targeting Env gene products. T20 is a synthetic peptide of 36 amino acids derived from a CHR fragment of B subtype HIV-1 gp41, which competes for binding of the NHR and thus blocks the formation of the hairpin structure that brings the virus and cell membranes into proximity for fusion. The functional relevance and utility of conservation of the gp41 region mimicked by C40 is illustrated in the broad recognition of C40 as an antigen by pooled plasma from 12 patients. The authors propose that a collection of such mimotopes can provide effective antigen-like characteristics for vaccine development.
In conclusion, the results suggest that neutralizing responses directed at epitopes like the gp41 CHR epitope may protect SPs through reasonably potent inhibition of Env-mediated cell fusion. The results of this study advance the generation of reliable and high-yield mimotope collections of HIV epitopes. Sets of epitopes recognized by HIV-1-specific NAbs provide useful probes for further analysis into the mechanistic details of cell fusion, and form a basis for deriving therapeutically useful antibody-based HIV-1 fusion inhibitors.
Conflict of interestAll authors declare to have no conflict of interest.
The authors are grateful to the patients who participated in this study. They would like to thank the doctors from Henan CDCs for their assistance. This work is supported by Mega-projects of Science Research for the 12th Five-Year Plan (2012ZX10001-006), Mega-projects of Science Research for the 11th Five-Year Plan (2008ZX10001-012), and National Natural Science Funds (30901274).