Carbapenemases have great importance in the global epidemiological scenario since infections with carbapenemase-producing bacteria are associated with high mortality, especially in hospitalized patients in intensive care units. This study describes two microorganisms producers of the New Delhi Metallo-b-lactamase, Klebsiella pneumoniae and Citrobacter freundii, from two patients admitted to a public hospital in Salvador, Bahia. These are the first clinical cases of New Delhi Metallo-b-lactamase described in microorganisms in the north and northeast Brazil. The isolates were characterized by antimicrobial susceptibility test, with resistance to all β-lactams including carbapenems, negative Modified Hodge Test and the synergy test with Ethylenediaminetetraacetic acid, Phenylboronic Acid and Cloxacillin was positive only with Ethylenediaminetetraacetic acid (difference of >5mm in the inhibition zone between the disk without and with the inhibitor). Analysis by multiplex PCR for blaIMP, blaVIM, blaNDM, blaKPC and blaOXA-48 enzymes confirmed the presence of blaNDM gene. This report of two different New Delhi Metallo-b-lactamase-producing microorganisms in a different region of Brazil confirms the risk of spreading resistance genes between different species and emphasizes the need for prevention and control of infections caused by these pathogens, which have limited treatment options and have been linked to high mortality rates.
Carbapenemases have great importance in the global epidemiological scenario since infections with carbapenemase-producing bacteria are associated with high mortality, especially in hospitalized patients in intensive care units. In addition, there are few treatment options available, coupled with the potential for transmission of resistance to other species through mobile genetic elements.1
To date, research has shown that resistance to β-lactam antibiotics in Enterobacteriaceae could be mediated by different β-lactamases, including those that degrade carbapenems, called carbapenemases. Further, the Metallo-β-lactamase (MBLs) are classified as Class B of Ambler and differ from other carbapenemases by using zinc at the active site, which facilitates the hydrolysis of the antibiotic.2
In 2009, a group of researchers from India described a new variant of MBL, called New Delhi Metallo-β-lactamase (NDM), encoded by the gene blaNDM.3 The NDM is endemic in the Indian subcontinent and since its first description it has been reported in cases of infections worldwide, often associated with international travel, medical tourism and potential exposures in the Balkans and the Indian subcontinent.4 By the year 2012, there were no reports of microorganisms producing this enzyme in South America and Antarctica.5,6 The first description of NDM in Brazil was in Providencia rettgeri and Enterobacter hormaechei isolated from Rio Grande do Sul State in 2013. After the first report an active surveillance was initiated, and there have been other reports involving NDM-producing Enterobacteriaceae, such as Enterobacter cloacae, Morganella morganii, Escherichia coli, and Klebsiella pneumoniae.7
These are the first clinical cases of NDM described in microorganisms derived from samples from two patients admitted to the Federal University Hospital in Salvador, State of Bahia. These are the first cases of NDM described in the north and northeast Brazil.
Patient AThe first patient was a 25-year-old man admitted to the oncohematological ward at the hospital on July 17th 2015. He had a previous diagnosis of xeroderma pigmentosum since childhood and was diagnosed with pancytopenia three months before admission due to episodes of diffuse bleeding (epistaxis, ear and gingival bleeding). He also had infected ulcers at the left leg. Previously he was admitted to another hospital, where he was transfused, treated for febrile neutropenia and diagnosed with bone marrow aplasia in a bone marrow aspirate in May 25th 2015, and then transferred to our hospital. During the hospitalization he was diagnosed with acute myeloid leukemia on July 23rd 2015, and chemotherapy was initiated. During treatment, he evolved with febrile neutropenia and blood cultures as well as culture of the catheter were drawn in August 8th 2015. The same carbapenem resistant strain of K. pneumoniae grew from these samples and were investigated for the presence of carbapenemase (described below). Antibiotic therapy was introduced with polymyxin and tigecycline, but the patient died on day 40 of hospitalization.
Patient BThe second patient was a 75-year-old man with previous diagnosis of systemic arterial hypertension, admitted to our hospital on July 28th 2015 due to a 15-day history of progressive muscular weakness in lower limbs, associated with urinary retention. Prior to admission in our hospital he was diagnosed with Guillain-Barrè Syndrome in another hospital where the patient was treated with immunoglobulin and ceftriaxone. The patient was then transferred to our hospital using urinary catheter, which was withdrawn at admission. Blood and urinary cultures from admission day were negative. During hospitalization, he evolved with respiratory distress being moved to the intensive care unit (ICU) of the hospital, but was discharged 4 days later with no need of invasive ventilatory support. Seven days after ICU discharge he presented with fever and the urinary culture collected four days later (on August 13th 2015) grew Citrobacter freundii resistant to all carbapenems, and was also investigated for the presence of Carbapenemase. This culture was interpreted as colonization, and the patient did not present any other episode of fever.
The identification and antimicrobial susceptibility tests of the bacteria isolated from the two patients were performed on Vitek 2 (bioMerieux – Marcy I’Etoile, France) and interpreted in accordance with the parameters of the Clinical and Laboratory Standards Institute.8 Both K. pneumoniae isolates showed resistance to all β-lactams including carbapenems, aminoglycosides and fluoroquinolones. The isolated C. freundii showed resistance to all β-lactams including carbapenems, but retained in vitro sensitivity to aminoglycosides and fluoroquinolones. The minimum inhibitory concentrations (MICs) to carbapenems were confirmed using Etest (bioMerieux – Marcy I’Etoile, France). The three isolates analyzed showed MICs to ertapenem, meropenem and imipenem higher than 32μg/mL.
After confirming the resistance to carbapenems, Modified Hodge Test (MHT) and phenotypic testing discs synergy with meropenem, imipenem, ertapenem and with and without Ethylenediaminetetraacetic acid (EDTA) 0.1M, cloxacillin and phenylboronic acid were performed. The MHT were negative and synergy tests showed increased inhibition zone (>5mm) in carbapenems discs in the presence of EDTA, EDTA-free in relation to the discs, and were negative with phenylboronic acid and cloxacillin for the microorganisms tested.
The isolates were then analyzed by multiplex PCR for blaIMP, blaVIM, blaNDM, blaKPC and blaOXA-48 enzymes, as described below.9
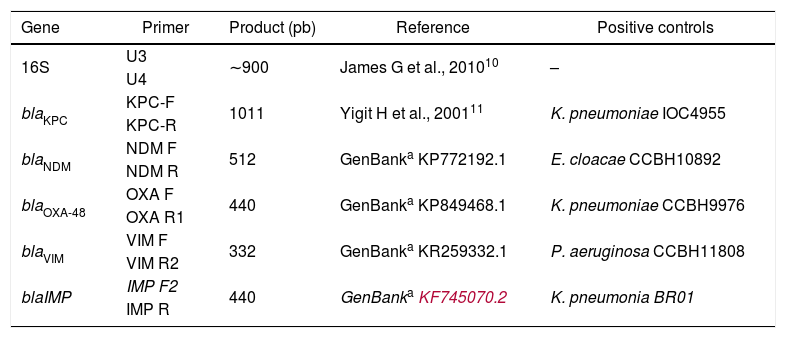
The bacterial lysates were prepared by suspending the colony in 500mL of deionized water (Milli-Q system, Millipore, Bedford, MA) and subjected to boiling for 15min and then frozen. The K. pneumoniae ATCC 700603 strain was used as a negative control in the analyses. Each PCR tube contained 20μL of the reaction mixture. PCR was performed using 2X READYMIX KappaTaq containing DNA polymerase (0.05U/uL, 1.25U per 25μL) reaction buffer with Mg2+ and 0.4mM of each dNTP, with loading dye (KappaBiosystems, Japan), primers (Table 1) and 2μL of bacterial lysates. All reactions were subjected to reaction control amplification using 16S gene.
The multiplex PCR (carbapenemase and 16S genes) by using specific oligonucleotide primers.
| Gene | Primer | Product (pb) | Reference | Positive controls |
|---|---|---|---|---|
| 16S | U3 | ∼900 | James G et al., 201010 | – |
| U4 | ||||
| blaKPC | KPC-F | 1011 | Yigit H et al., 200111 | K. pneumoniae IOC4955 |
| KPC-R | ||||
| blaNDM | NDM F | 512 | GenBanka KP772192.1 | E. cloacae CCBH10892 |
| NDM R | ||||
| blaOXA-48 | OXA F | 440 | GenBanka KP849468.1 | K. pneumoniae CCBH9976 |
| OXA R1 | ||||
| blaVIM | VIM F | 332 | GenBanka KR259332.1 | P. aeruginosa CCBH11808 |
| VIM R2 | ||||
| blaIMP | IMP F2 | 440 | GenBankaKF745070.2 | K. pneumonia BR01 |
| IMP R |
The solutions were subjected to the following cycling conditions: initial denaturation of 95°C (2min, 1 cycle); followed by 40 cycles: denaturation 95°C (30s), annealing 55°C or 57°C (30s) and extension 72°C (2min, 1 cycle); and a final extension cycle of 72°C (2min). We used the gradient thermal cycler Mastercycler® System, Eppendorf, Germany. The PCR product (amplicon using 15μL in gel) were visualized after electrophoresis (2% agarose), and stained with ethidium bromide (10mg/mL) in Kodak documentation system (Gel Logic 100 Imaging System).
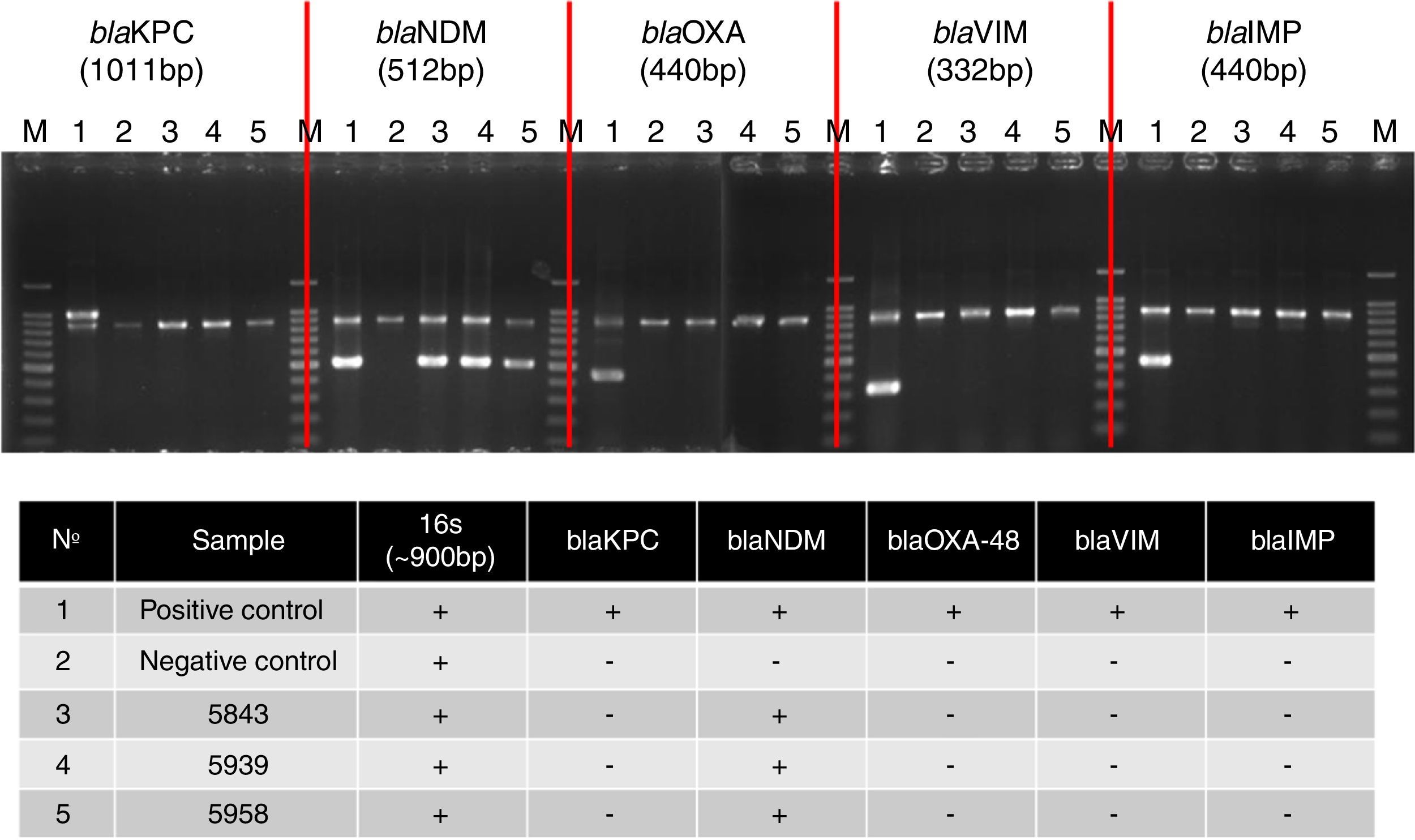
Fig. 1 shows banding patterns of multiplex PCR reactions for the genes studied. The blaNDM gene was amplified, confirming the enzymatic profile of microorganisms.
Multiplex PCR assay for simultaneous detection of 16S rRNA (∼900bp), as an amplification control, and carbapenemase genes. BlaKPC: Lane 1, K. pneumonia IOC4955 (positive control); Lane 2, K. pneumonia ATCC 700603 (negative control); Lane 3, 5843; Lane 4, 5939; Lane 5 5958. BlaNDM: Lane 1, E. cloacae CCBH10892 (positive control); Lane 2, K. pneumonia ATCC 700603 (negative control); Lane 3, 5843; Lane 4, 5939; Lane 5 5958. BlaOXA-48: Lane 1, K. pneumoniae CCBH9976 (positive control); Lane 2, K. pneumonia ATCC 700603 (negative control); Lane 3, 5843; Lane 4, 5939; Lane 5 5958. BlaVIM: Lane 1, P. aeruginosa CCBH11808 (positive control); Lane 2, K. pneumonia ATCC 700603 (negative control); Lane 3, 5843; Lane 4, 5939; Lane 5 5958. BlaIMP: Lane 1, K. pneumonia BR01 (positive control); Lane 2, K. pneumonia ATCC 700603 (negative control); Lane 3, 5843; Lane 4, 5939; Lane 5 5958. Lane M, 100-bp DNA ladder (Invitrogen). The electrophoresis was run in a 2% agarose gel, which was stained with ethidium bromide.
Enterobacteria that carry resistance gene to carbapenems, including blaNDM, are considered an important public health problem because of the possibility of expansion of the genes and the clinical impact on the management of infections associated with these microorganisms, which are typically resistant to almost all antibiotics.12
After the first NDM isolate in 2009 in India, the NDM has been detected in every continent, including Brazil. In South America, blaNDM gene was first reported in 2012 in Uruguay, a country that borders the Rio Grande do Sul state, where NDM was first described in Brazil.13 In addition, in both accounts, the gene was isolated from P. rettgeri. Since then, other NDM-producing Enterobacteriaceae (M. morganii, E. cloacae complex) were isolated in this state from clinical and surveillance cultures in different hospitals.14 More recently, it was first isolated from a surveillance swab, the blaNDM-gene in K. pneumoniae, in Rio de Janeiro. This isolate carried a resistance gene already found in a KPC strain in 2010 in the state of Minas Gerais, highlighting the ability of microorganisms to acquire and disseminate such resistance genes.15
This report of NDM isolates in two different microorganisms, K. pneumoniae and C. freundii, confirms the risk of spread of resistance genes between different species. The rapid spread of genes conferring resistance to carbapenems, blaKPC, blaNDM, among others, is of great concern because the emergence of multi-resistant microorganisms is a threat to public health due to the shortage of new antibiotics in development.16 This report emphasizes the need for strict measures to prevent and control infections, which can minimize the spread of genes conferring resistance to carbapenems among different species of Enterobacteriaceae.
Conflicts of interestThe authors declare no conflicts of interest.