Community-Acquired Pneumonia (CAP) is the primary cause of hospitalization in the United States and the third leading cause of death in Brazil. The gold standard for diagnosing the etiology of CAP includes blood culture, Gram-stained sputum, and sputum culture. However, these methods have low sensitivity. No studies investigating the etiology of CAP have been conducted in Brazil in the last 20-years, and the empirical choice of antimicrobials is mainly based on the IDSA guidelines. This is the first national study with this aim, and as a result, there's potential for the Brazilian consensus to be impacted and possibly modify its guidelines rather than adhering strictly to the IDSA's recommendations.
MethodsThe aim of this study is to identify the main microorganisms implicated in CAP by employing a multiplex Polymerase Chain Reaction (mPCR) at the foremost public hospital in Brazil. All patients who were admitted to the emergency department and diagnosed with severe CAP underwent an mPCR panel using nasopharyngeal and oropharyngeal swabs, with the aim of detecting 13 bacterial and 21 viral pathogens.
ResultsA total of 169 patients were enrolled in the study. The mPCR panel identified an etiological agent in 61.5% of patients, with viruses being the most common (42.01%), led by Rhinovirus, followed by Influenza and Coronavirus (non-SARS-CoV-2). Bacterial agents were identified in 34.91% of patients, with S. pneumoniae being the most common, followed by H. influenzae, M. catarrhalis, and S. aureus. Additionally, we found that the prescription for 92.3% of patients could be modified, with most changes involving de-escalation of antibiotics and antiviral therapy.
ConclusionOur study revealed different etiological causes of CAP than those suggested by the Brazilian guidelines. Using molecular diagnostic tests, we were able to optimize treatment by using fewer antibiotics.
Lower respiratory infections are one of the top ten causes of death according to the World Health Organization (WHO), and Community-Acquired Pneumonia (CAP) is the main cause of hospitalization in the United States, with medical costs exceeding $10 billion in 2011. CAP is also the third leading cause of death in Brazil.1-4
Traditionally, clinicians have used X-Rays in combination with clinical symptoms as a standard tool to diagnose pneumonia, however, several other causes can lead to the same findings. Additionally, until recent years, the gold standard for diagnosing the etiology of pneumonia was blood culture, Gram-stained sputum, and sputum culture. However, these methods have low sensitivity, and more than 50% of patients either do not produce sputum or have sputum of poor quality for analysis.5-11
The guideline of Infectious Diseases Society of America (IDSA) is commonly used to guide the diagnosis and treatment of CAP worldwide. Despite limited evidence, the guideline do not recommend using etiological diagnostic tests for CAP except in severe condition and maintain the same bacterial agents over the last decade (S. pneumoniae, H. influenzae, M. pneumoniae, S. aureus, Legionella spp, C. pneumoniae, and M. catarrhalis) as the main CAP pathogens. Nevertheless, a recent study conducted in the United States drew parallels between the etiological agents documented and those delineated in its respective guideline, revealing a concordance rate of under 30%, the agreement will be even less when another country, such as Brazil, uses them as a foundation for its own guidelines.12-15
The Polymerase Chain Reaction (PCR) has been shown to be more sensitive than traditional diagnostic methods for determining the etiology of CAP.16,17 Recent studies using this method have identified different etiologies than those suggested by the guidelines, including respiratory viruses.3,18
No studies investigating the etiology of CAP have been conducted in Brazil in the last 20-years, and the empirical choice of antimicrobials is mainly based on the IDSA guidelines. The aim of this study is to use a multiplex Polymerase Chain Reaction (mPCR) panel in conjunction with traditional diagnostic methods (blood culture, Gram-stained sputum, and sputum culture) to identify the main microorganisms responsible for community-acquired pneumonia at the largest public hospital in Brazil.
MethodsStudy designFrom September 2017 to August 2018, we screened all patients admitted to the emergency room of Hospital das Clínicas da Faculdade de Medicina da Universidade de Sao Paulo (ER-HC-FMUSP) who required hospitalization based on clinician decision. ER-HC-FMUSP is a teaching hospital that serves as a reference for all severe clinical and surgical cases. During this period, there were 6,205 clinical admissions. We attempted to enroll all eligible adults between Sunday and Thursday over the course of one year in order to include all seasonal pathogens. Due to laboratory operations, enrollment was limited to these days. Written informed consent was obtained from all patients or their caregivers prior to enrollment
Patients were enrolled if they were older than 18-years, had evidence of CAP defined as lung imaging (chest radiography or computed tomography), and two of the following symptoms: fever (temperature >37.8°C), cough, sputum production, shortness of breath, pleuritic chest pain, mental confusion, leukocytosis (White Blood Count [WBC] >12,000 mm3), or a suppressed WBC count (<6,000 mm3).
Patients were excluded if they had been recently hospitalized (<30 days), admitted more than 48 hours before enrollment, undergone hematopoietic stem cell transplantation, received chemotherapy in the past 30 days, received antibiotics for over 48 hours before enrollment, were postoperative, or had a clear alternative diagnosis.
Severity was evaluated using the Pneumonia Severity Index (PSI) Score.19
Antibiotic appropriateness was verified using the “Sanford Guide Antimicrobial Stewardship” book.20
The primary outcome was to identify the main microorganisms responsible for community-acquired pneumonia at the largest public hospital in Brazil. The secondary outcome involves assessing diagnostic gain using the mPCR methodology compared to standard methodology and the appropriateness of the prescribed antibiotic.
Specimen collectionBlood cultures, sputum (if patients had productive cough), nasopharyngeal and oropharyngeal swabs were obtained from all patients after obtaining their consent. The results of the PCR tests were not available until the end of the study and were not used for patient care.
Laboratory testingBlood and high-quality sputum samples were collected using standard techniques.
The mPCR panel was performed on nasopharyngeal and oropharyngeal swabs. For the first 107 patients, the Mobius Life Science's kit was used, which identified the following pathogens: Influenza A; Influenza B; Influenza C; Influenza A (H1N1); Parainfluenza viruses 1, 2, 3 and 4; Coronaviruses NL63, 229E, OC43 and HKU1; Human metapneumoviruses A/B; Rhinovirus; Respiratory syncytial viruses A and B; Adenovirus; Enterovirus; Parechovirus; Bocavirus; Pneumocystis jirovecii; Mycoplasma pneumoniae; Chlamydophila pneumoniae; Streptococcus pneumoniae; Haemophilus influenzae; Haemophilus influenzae type B; Staphylococcus aureus; Moraxella catarrhalis; Bordetella spp.; Klebsiella pneumoniae; Legionella pneumophila; Legionella longbeachae and Salmonella spp . However, due to the discontinuation of the Mobius Life Science's kit in the hospital laboratory, the Biomerieux's Biofire FilmArray kit was used for the last 57 patients. This kit identified: Influenza A; Influenza B; Influenza A (H1N1); Parainfluenza viruses 1, 2, 3 and 4; Coronaviruses NL63, 229E, OC43 and HKU1; Human metapneumoviruses A/B; Rhinovirus/Enterovirus; Respiratory syncytial viruses A and B; Adenovirus; Mycoplasma pneumoniae; Chlamydia pneumoniae; Bordetella parapertussis and Bordetella pertussis.
Statistical analysis and ethicsAll data was stored on the Redcap platform. We used prevalence, mean, and standard deviation for parametric data, and median and interquartile range (p25%‒p75%) for non-parametric data. The Shapiro-Wilk test was used to determine if the data was normally distributed, with statistical significance set at a p-value of <0.05. The analyses were performed using Stata 15.1 software.
For the Staphylococcus aureus bacteria results, considering its potential ability to colonize the upper airways and skin, and the fact that carriage has been reported to be highly variable between populations (4%–64%),21 we used a lowered threshold cycle of interquartile p25% to consider a positive test.
The study was approved by the Ethics Committee at HC-FMUSP (CAPPesq 11751), and written informed consent was obtained from all participants. The principles of the Declaration of Helsinki were followed, and the study was conducted according to good clinical practice guidelines.
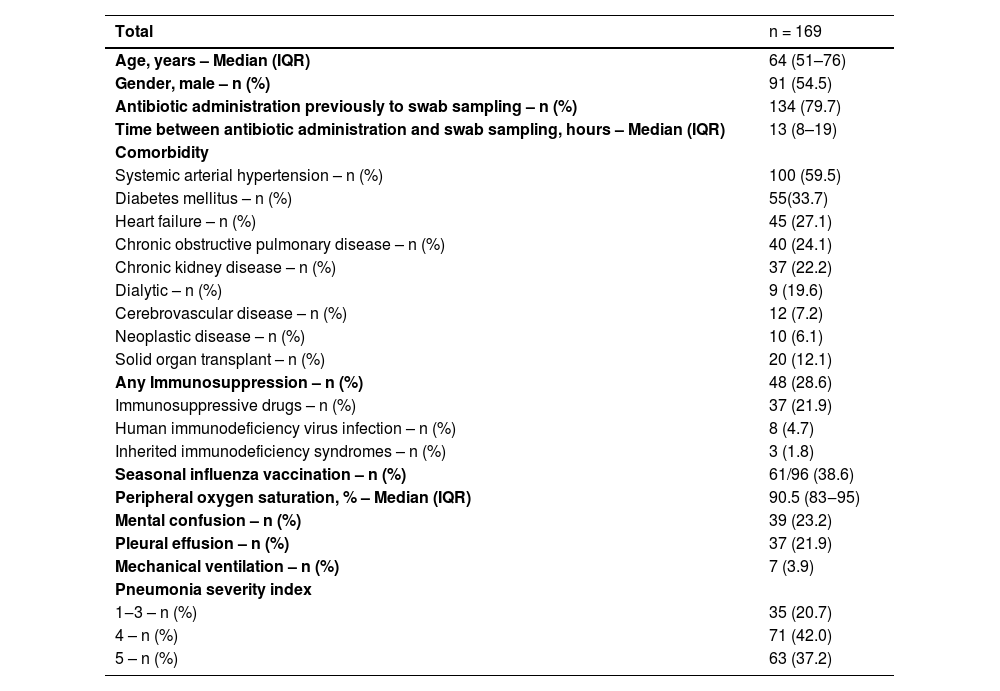
ResultsOut of 172 eligible patients, 169 were enrolled in the study. The mean age was 64 years and 54% of them were male. The most common comorbidity was hypertension, followed by diabetes mellitus, heart failure, and chronic obstructive pulmonary disease. About 28.6% of the patients had some immunosuppressive condition. Seven patients required intubation upon arrival at the emergency room (Table 1).
Baseline characteristics of patients with community-acquired pneumonia requiring hospitalization at the ER-HC-FMUSP.
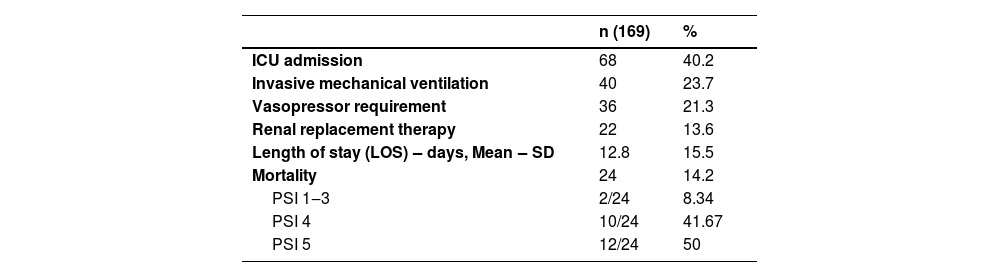
Among the 169 patients, 40% required admission to the intensive care unit, 23% progressed to respiratory support with invasive mechanical ventilation, and 13.6% required renal replacement therapy. The mean length of hospital stay was 12.8-days, and the mortality rate was 14.2% (Table 2).
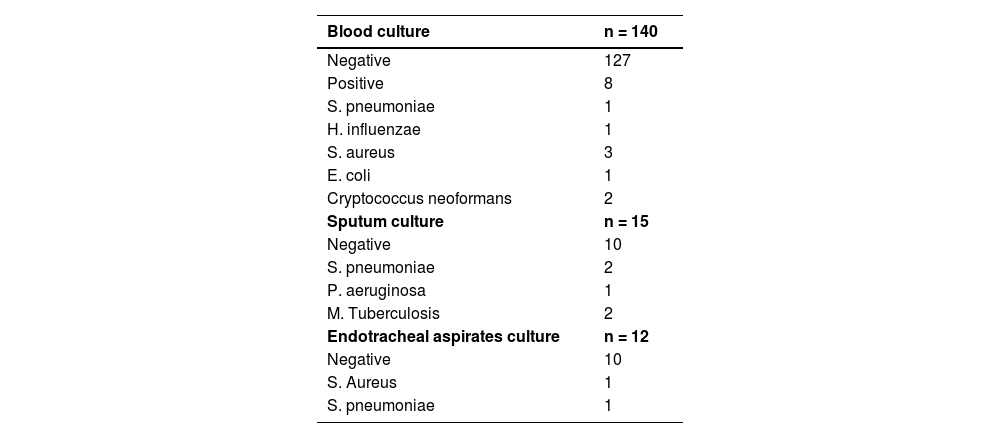
The blood culture has been collected from 140 patients with only 5.7% of them with positive's results. Sputum cultures and endotracheal aspirate cultures were collected from 15 and 12 patients, respectively. The results are described in Table 3. When we analyzed the positive cases using the standard etiology investigation, we found a total of 11 cases, representing 6.52% of the total number of patients.
The mPCR panel identified an etiological agent in 61.5% of patients, with viruses being the most common (42.01%), led by Rhinovirus, followed by Influenza and Coronavirus (non-SARS-CoV-2). Bacterial agents were identified in 34.91% of patients, with S. pneumoniae being the most common, followed by H. influenzae, M. catarrhalis, and S. aureus (Table 4).
Pathogen detection in patients with community-acquired pneumonia molecular methods (n = 169).
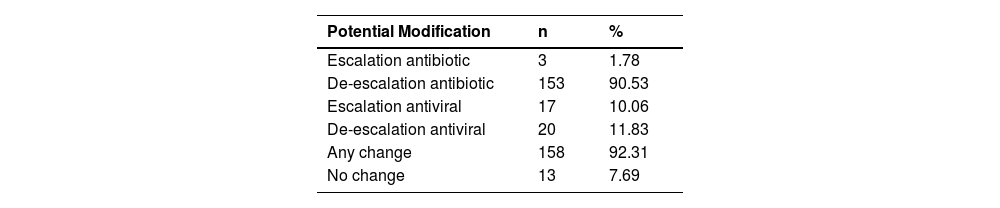
Adopting both diagnostic methods, we found that the prescription for 92.3% of patients could be modified, with most changes involving de-escalation of antibiotics and antiviral therapy (Table 5). The primary class of antibiotics that could be subject to modification was macrolides.
DiscussionTraditional methods of etiological diagnosis for Community-Acquired Pneumonia (CAP) have been restricted to the investigation of bacterial agents and have limited sensitivity.5-9 These limitations have led ISDA and Brazilian consensus guidelines to not recommend etiological investigation for mild cases or in patients without comorbidities. Even for cases requiring hospitalization, obtaining respiratory samples for identification of the agent is only recommended for those with severe forms of the disease or at risk of infection with resistant bacteria.14 These recommendations are justified by their lack of impact on clinical outcomes in addition to the cost of the tests. We observed a low frequency of collection of sputum or tracheal secretion samples, demonstrating the difficulty in obtaining these samples. In addition, despite the high frequency of blood culture collection, the positivity rate was quite low, bringing little contribution to guide the antimicrobial. These factors contribute to the recommendation of empirical use of antimicrobials in these guidelines, especially with coverage against pneumococci, as coinfection with viral and bacterial agents is common.12,14,22-25 This situation is further exacerbated in Brazil, where there is a lack of comprehensive knowledge regarding the primary causative agents of CAP. Consequently, the American guidelines are employed as a reference, both for determining etiological agents and for the treatment of CAP in our hospital.
However, the increasing antimicrobial resistance is becoming a global problem, including for community infections, to the point that in 2001, the World Health Organization drew attention to this scenario and outlined strategies to contain this problem, which has as its first step the rational use of antimicrobials.26
The H1N1 influenza pandemic in 2009 reinforced the possibility of a viral agent causing severe acute respiratory failure and ARDS, including radiological consolidation patterns and the potential for fatal outcomes.27 This scenario was further reinforced during the COVID-19 pandemic, where several authors observed that the isolated viral agent could be responsible for indistinguishable cases of bacterial pneumonia, even in patients admitted to the ICU.28-30
Our study shows that expanded etiology investigation for CAP using molecular diagnostic methods can increase pathogen detection from 6.52% to 61.54%, representing a 55% increase in positive diagnoses. With this more sensitive approach, we described a different prevalence of the principal microorganisms causing severe CAP in our country. We are the first study to investigate the etiology of severe CAP in Brazil, and we found different microorganisms compared to those described in national guidelines.25 The most prevalent agent was a virus (61.54%), while bacteria were found in only 34.91% of the samples, being S. pneumoniae more prevalent followed by, H. influenzae, M. catarrhalis, and S. aureus. These results are similar to studies using molecular diagnostic tests in other countries.3
Another contribution of our study is the low prevalence of M. pneumoniae. In the Brazilian guideline for treating CAP, it's mentioned as the second most prevalent agent, but our findings indicate an incidence of only 1.78%. Although consensus recommends introducing a macrolide as antibiotic therapy, we observed that this may not be necessary as a routine. Our results are consistent with recent studies worldwide that use this molecular technique.3,17,18
Upon estimating the potential impact of molecular testing on the treatment of patients diagnosed CAP, we observed a significant increase in diagnostic accuracy, with the percentage rising from 6.52% to 61.5%, representing a nearly 55% rise as compared to the standard methodology.
The indiscriminate use of antibiotics in clinical practice is a concern, and in our study, we found that using a molecular diagnostic test for CAP could lead to more than 90% of treatments being changed, resulting in a significant de-escalation in antibiotic use.
A limitation of our study is that it was conducted in only one hospital and therefore cannot be extrapolated to the entire country. Further studies need to be done to confirm our hypothesis. Another limitation is that we used a nasopharyngeal swab for etiological investigation, which means that some agents may be just colonization and not the source of infection. However, as we only evaluated patients who presented clinical symptoms compatible with pneumonia and signs of severity by the PSI score, we consider that if we isolate an etiological agent that is plausible to cause infection, it should be considered as the responsible agent for the infection and not just colonization. Another limitation of our study is that patients were already using antibiotics at the time of collecting respiratory samples. This could potentially have influenced the negativity of the culture tests and PCR panel results.
In conclusion, despite our limitations, we found different etiological causes of PAC than those suggested by the Brazilian guidelines. Using molecular diagnostic tests, we were able to optimize our treatment by using fewer antibiotics.