A clinical Providencia stuartii isolate SM662 was recovered from a patient hospitalized in the intensive care unit at the Military hospital, Tunisia. This isolate was resistant to penicillins, cephalosporins, aminoglycosides and fluoroquinolones. A marked in vitro synergy between ceftazidime or cefotaxime and amoxicillin–clavulanic acid on Mueller-Hinton agar plates suggested the presence of an extended-spectrum-β-lactamase. In addition, an unusual synergy was found between cefepime or aztreonam, and cefoxitin or imipenem on a double disk synergy test suggesting a VEB-type extended-spectrum-β-lactamase. The characterization of β-lactamases and associated resistance genes was performed by isoelectric focusing, polymerase chain reaction and nucleotide sequencing. Two β-lactamases bands with pI values of 5.4 and 7.7, which were matched to TEM-1, VEB-1-a and OXA-2-like β-lactamases were detected. The blaVEB-1-a gene was found to be associated with complex genetic structures, including Re elements. These β-lactamases were not transferred by electroporation or conjugation experiments to the transconjugants and electroporants. Hybridization methods showed that the extended-spectrum-β-lactamase encoding gene may have a chromosomal localization. The isolate SM662 produced the quinolone resistance determinants qnrA6 and aac(6′)-Ib-cr which were successfully transferred. The detection of an associated chromosomal quinolone resistance revealed the presence of a gyrA mutation at codon 83 (Ser83Ile). This is the first report of the linkage VEB-1-a/OXA-2-like in P. stuartii associated with the description of qnrA6 and aac(6′)-Ib-cr genes in this isolate.
Providencia stuartii is a frequent cause of urinary tract infections in hospitalized patients.1 It plays an important role as a nosocomial pathogen in the dissemination of plasmid-mediated resistance.2P. stuartii is naturally resistant to aminopenicillins and narrow-spectrum cephalosporins due to a chromosomally expressed Ambler class C cephalosporinases (AmpC).1 However, acquisition of ESBL has been reported.1,2blaVEB-1 gene was identified in P. stuartii for the first time in Alger.1 VEB-1 β-lactamase confers high-level resistance to a broad spectrum of cephalosporins; however this activity is inhibited not only by clavulanate, but also by cefoxitin and imipenem.3 Until now, the blaOXA-2 gene has not been detected in Providencia genus as to the best of our knowledge, but in Tunisia it was described in clinical strains of Pseudomonas aeuroginosa.4 A decreased quinolone susceptibility associated with qnrA6 and aac(6′)-Ib-cr determinants was also reported in Tunisia in clinical strains of P. stuartii.5 In the current study, we report for the first time the co-production of chromosomal blaVEB-1-a and blaOXA-2-like genes in a multidrug resistant P. stuartii clinical strain isolated at the Military hospital in Tunisia and their association with plasmid-mediated qnrA6 and aac(6′)-Ib-cr determinants.
On July 2008, a 46-year-old man was transferred from a Tunisian Regional Hospital and he was hospitalized in the intensive care unit at the Military hospital in Tunis, Tunisia for a severe cranial traumatism. Three months thereafter, the patient notably diabetic and epileptic was febrile at any time and subsequently developed a chronic infection. According to the patient's medical records, such an infection turned out to be urinary tract infection that was diagnosed following the appearance of an infectious syndrome; it was treated with ceftazidime and ofloxacin. At the end of October 2008, P. stuartii SM662 isolate was recovered by a tracheal aspirate, although seven days prior to the isolation of this strain he had received a course of cefotaxime and ciprofloxacin. The patient was treated with gentamicin and imipenem. Seven days after starting antimicrobial therapy the clinical outcome indicated treatment failure and the ultimately died.
The P. stuartii SM662 strain was identified using an AP20E kit (Biomèrieux, Marcy-l’Etoile, France). E. coli DH10B (Invitrogen, Life Technologies) and streptomycin resistant E. coli HB101 recipient strains were used respectively for the electroporation and conjugation experiments. β-Lactamases with known pIs were used as standards: TEM-1 (pI 5.4), TEM-2 (pI 5.6), TEM-3 (pI 6.3) and SHV-1 (pI 7.6).6 Antimicrobial susceptibility was determined by the disk diffusion method on Mueller-Hinton (MH) agar (Bio-Rad, Marnes La Coquette, France) recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines.7 The isolate was resistant to multiple antibiotics, including chloramphenicol, kanamycin, tobramycin, sulphonamide, tetracycline, nalidixic acid, ciprofloxacin and ofloxacin whereas it was susceptible to imipenem, ertapenem, gentamicin and piperacillin. The double disc synergy test was positive showing a marked synergy between ceftazidime or cefotaxime and amoxicillin–clavulanic acid on MH agar plates and suggested the presence of a class A ESBL.8 In addition, an unusual synergy was found between cefepime or aztreonam, and cefoxitin or imipenem on a double disk synergy test suggesting a VEB-type ESBL production according to Naas et al.3 The minimum inhibitory concentrations (MICs) (Table 1) were determined by the broth microdilution method and interpreted according to the CLSI criteria.6 The strain was found intermediately resistant to imipenem according to the novel CLSI breakpoints (M100-S23) and the difference on the carbapenem's activity (imipenem and ertapenem) is due to the lower activity of imipenem against Providencia spp., Proteus spp. and Morganella morganii.
MICs (μg/mL) of various antimicrobial agents obtained for the clinical isolate P. stuartii SM662, its transconjugants and electroporants and the E. coli HB101 and E. coli DH10B recipients strains.
| P. stuartii SM662 | HB101×SM662 | E. coli HB101 | DH10B/pSM662 | E. coli DH10B | |
|---|---|---|---|---|---|
| Amoxicillin | >512 | 4 | 8 | 4 | 2 |
| Ticarcillin | >512 | 4 | 8 | 8 | 2 |
| Oxacillin | 512 | <4 | <2 | 2 | 2 |
| Cefoxitin | 64 | 8 | 2 | 4 | 4 |
| Cefotaxime | >512 | <4 | <2 | 2 | 4 |
| Ceftriaxone | 512 | <4 | <2 | 2 | 1 |
| Ceftazidime | >512 | <4 | <2 | 2 | 1 |
| Aztreonam | 512 | 4 | <2 | <4 | 1 |
| Nalidixic acid | >512 | >512 | 2 | >512 | 2 |
| Chloramphenicol | 256 | 8 | <2 | 8 | 2 |
| Tetracycline | 512 | 8 | <2 | 8 | <1 |
| Ciprofloxacin | 64 | 4 | 2 | 8 | 1 |
| Ofloxacin | 128 | 64 | 2 | 32 | 1 |
| Streptomycin | 64 | 256 | 512 | 128 | 1 |
| Tobramycin | 128 | 256 | <0.25 | 128 | 2 |
| Gentamicin | 1 | 0.5 | <0.25 | 0.5 | <0.25 |
| Piperacillin | 1 | 0.5 | <0.25 | 0.5 | <0.25 |
| Ertapenem | 0.25 | <0.25 | <2 | <0.25 | <0.25 |
| Imipenem | 2 | <2 | <2 | <2 | <1 |
Whole-cell DNA from P. stuartii SM662 was used as a template for PCR assays. Presence of blaTEM-1, blaVEB-1-a and blaOXA-2-like genes was assessed by PCR and sequencing as previously described.6,9,10 No amplicons were obtained with blaSHV and blaCTX-M genes.6,11 Furthermore, multiplex PCR amplifications using primers specific for plasmid-mediated AmpC β-lactamases (CBLs)12 were negative.
VEBcas-F and VEBcas-B (Eurogentec, Belgium) located at each end of the blaVEB-1 cassette were used to amplify the entire blaVEB-1 gene.3 Conditions PCR amplification experiments were performed using primers located in the blaVEB-1a gene and in the class 1 integron variable region (5′CS–3′CS) (Eurogentec, Belgium) as described previously.13 Amplification of the class 1 integron variable region (5′CS–3′CS) was positive in SM662 showing size of about 1200bp. Sequence analysis showed two genes cassettes arrays: aadB+dfrA1. A combination of 5′-CS or 3′-CS primers and VEBINV1 or VEBINV2 (Eurogentec, Belgium), respectively, both primers reading outwards from blaVEB-1, was also used for the determination of the genetic content of class 1 integron.13 However, no PCR fragments were obtained suggesting that the blaVEB-1a gene cannot be part of a class 1 integron. This hypothesis did not guarantee that this gene was not inserted into a class 1 integron, since VEB-1 is usually described as part of a gene cassette itself located in a class 1 integron.3 A further PCR performed using primers pair Re1F (repeat element) and VEBcas-B,14 revealed the presence of a PCR fragment of about 1.2 Kb and suggested that the blaVEB-1a gene was associated to two Re1 repeated elements in the direct orientation. A previous report identified the presence of Re1 repeat elements sequences surrounding the blaVEB-1-a gene in P. aeruginosa 10.2 clinical isolate from India [14].
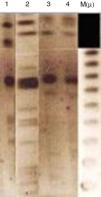
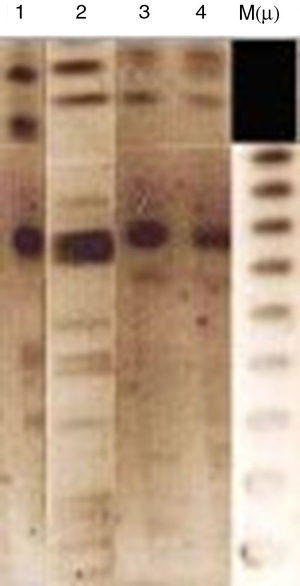
Analytical isoelectric focusing of crude β-lactamase extract of P. stuartii SM66215 demonstrated two bands of β-lactamases activities with pIs of 5.4 and 7.7. TEM-1 and VEB-1 have both 5.4 while OXA-2 has the pI 7.7. These β-lactamases have been not transferred suggesting that were not mediated by a conjugative or transferable plasmid. The single plasmid transferred (p-SM662) using a plasmid extraction kit GFX Micro Plasmid Prep (Amersham Biosciences, UK), conferred resistance only to nalidixic acid, ofloxacin and tobramycin (Table 1). Hybridization methods after digestion restriction with SmaI and HindIII (Biorad®, Laboratories, France)16 showed that blaVEB-1a and blaOXA-2 like genes may have a chromosomal localization (Fig. 1). Several studies reported that blaVEB-1-a like genes are mostly plasmid located in Enterobacteriaceae, whereas they are chromosomally located in P. aeruginosa and Acinetobacter baumannii.1 Nonetheless, in our study we identified a chromosomal VEB-1-a type ESBL. This finding is described for the first time in Tunisia and suggests that blaVEB-1-a can spread among clinically relevant species. Here, we describe a multidrug resistant P. stuartii SM662 co-produced TEM-1 and the narrow-spectrum β-lactamase OXA-2-like, together with the ESBL VEB-1-a. Previous finding reported the simultaneous presence of blaVEB-1 and blaoxa-10 genes in a clinical strain of P. stuartii V1 isolated from Nigeria17 but our study presents the first report of the linkage VEB-1-a/OXA-2-like in P. stuartii SM662 clinical isolate, to the best of our knowledge. Blaoxa-2-like has been detected in P. aeruginosa isolates from Tunisia;4 this finding indicated that OXA genes can spread progressively between species.
Interestingly, a marked association was found between ESBL production and multidrug resistance. To investigate the coresistance, PCR detection and sequencing of the qnrA, qnrB and qnrS genes18 and the aminoglycoside/fluoroquinolone-modifying enzyme-encoding aac(6′)-Ib-cr gene19 identified the qnrA6 determinant and the variant aac(6′)-Ib-cr on the same plasmid. The transconjugants and the electroporants expressed non-susceptibility to ofloxacin, streptomycin and tobramycin (Table 1). In our study, aac(6′)-Ib-cr, which encodes an aminoglycoside acetyltransferase, was found associated with VEB-1 and OXA-2 β-lactamases for the first time in clinical strain of P. stuartii in Tunisia. PCR detection and sequencing of an additional chromosomal quinolone resistance determinants regions (QRDRs)20 did not reveal the presence of gyrB, parC and parE, but we detect a gyrA mutation at codon 83 (Ser-Ile). This observation may explain the higher level of resistance to nalidixic acid in our isolate.
In conclusion, our study indicated for the first time in Tunisia the dissemination of VEB-1 β-lactamase associated with plasmid-mediated qnrA6 and aac(6′)-Ib-cr-like determinants in a multidrug resistant P. stuartii clinical isolate. The presence of Re sequences surrounding the blaVEB-1-a gene is worrying since their origin and their function in the mobilization of blaVEB-1-a remain unknown and pose a challenge for the treatment of hospital infections due to Gram-negative bacteria. Therefore, the incidence of ESBL-producing bacteria needs a continuous monitoring of such multidrug resistant strains and warrants further study of their epidemiologic evolution.
Conflicts of interestThe author declare no conflicts of interest.
The authors wish to thank Pr. Ferjani Mustafa, director of Intensive Care Unit War, Military hospital, Tunis for his helpful assistance to obtain clinical data of the isolate. This work was supported by the Tunisian Ministry of Higher Education, Scientific Research and Technology.