Response-guided therapy is of limited use in developing countries because hepatitis C virus RNA detection by sensitive molecular methods is time- and labor-consuming and expensive. We evaluated early predictive efficacy of serum hepatitis C virus core antigen kinetics on sustained virologic response in patients with genotype 1 hepatitis C virus during pegylated interferon plus ribavirin treatment. For 478 patients recruited, hepatitis C virus RNAs were detected at baseline, and at weeks 4, 12, 24, 48, and 72 using Cobas TaqMan. Architect hepatitis C virus core antigen was performed at baseline, and weeks 4 and 12. Predictive values of hepatitis C virus core antigen on sustained virologic response were compared to hepatitis C virus RNA. In the first 12 weeks after treatment initiation the dynamic patterns of serum hepatitis C virus core antigen and hepatitis C virus RNA levels were similar in sustained virologic response, relapse, and null response patients groups. Although areas under the receiver operating characteristics curves of hepatitis C virus core antigen were lower than those of hepatitis C virus RNA at the same time points, modeling analysis showed that undetectable hepatitis C virus core antigen (rapid virological response based on hepatitis C virus core antigen) had similar positive predictive value on sustained virologic response to hepatitis C virus RNA at week 4 (90.4% vs 93.3%), and hepatitis C virus core antigen decrease greater than 1log10IU/mL (early virological response based on hepatitis C virus core antigen) had similar negative predictive value to hepatitis C virus RNA at week 12 (94.1% vs 95.2%). Analysis on the validation group demonstrated a positive predictivevalue of 97.5% in rapid virological response based on hepatitis C virus core antigen and a negative predictive value of 100% in early virological response based on hepatitis C virus core antigen. In conclusion, hepatitis C virus core antigen is comparable to hepatitis C virus RNA in predicting sustained virologic response of chronic genotype 1 hepatitis C virus infected patients, and can be used to guide anti-hepatitis C virus treatment, especially in resource-limited areas.
It is estimated that more than 185 million people around the world have been infected with hepatitis C virus (HCV), and the South Asian and East Asian regions have by far the largest number of persons living with HCV infection, where more than 100 million people were infected by HCV.1 The burden of disease including the costs of treatment and monitoring are greater in developing countries.2
Pegylated interferon (PegIFN) plus ribavirin (RBV) was the standard of care for patients with chronic HCV infection before 2011.3 Following the appearance of direct acting antiviral (DAA) drugs when two protease inhibitors (telaprevir and boceprevir) were licensed for use as the first-generation DAAs, the field of anti-HCV treatment is evolving rapidly in chronic hepatitis C (CHC).4 However, the available data for the DAAs are from drug registration trials, and data on safety and long-term effectiveness are also limited because of the small number of persons included in the studies in addition to short follow-up. Most importantly, DAAs are very expensive; a 12-week course of sofosbuvir costs as much as US$ 84,000 in the US.5 Therefore, it is likely that PegIFN and RBV will be the only available regimen for the next several years in most countries. The WHO strongly recommends that PegIFN in combination with RBV be used for the treatment of chronic HCV infection, especially in resource-limited settings.5 Even the 2014 EASL Guidelines recommend PegIFN/RBV for selected patients with high likelihood of sustained virologic response (SVR).6
Even standard-of-care regimens have problems related to high cost of treatment, requirement for sophisticated laboratory testing before and during treatment, high rate of adverse events, and unsatisfactory virological response.6,7 It is very important to predict the virologic response in order to adjust the treatment protocol earlier. Response-guided therapy, in which HCV RNA quantification should be performed by a sensitive assay (lower limit of detection (LLOD) <50IU/mL, or even <15IU/mL), is widely used to guide the PegIFN/RBV dual therapy.6 However, in many low- and middle-income countries, sensitive HCV RNA assays are either not available or available only in some large cities.5 A simple and inexpensive surrogate marker is therefore required in clinical practice. It has been reported that HCV core antigen (HCVcAg) reflects viral replication, and its serum level varies following antiviral treatment in hepatitis C patients.8–10 Early kinetics of serum HCVcAg predicts SVR well in CHC patients on standard-of-care therapy, and HCVcAg level at week 1 is a stronger predictor of SVR.11,12 However, both studies had small sample sizes. Thus, further and larger studies were warranted in order to confirm the clinical significance of HCVcAg reduction during treatment.
Accordingly, based on phases IIb and III clinical trials that examined the safety and efficacy of type Y pegylated interferon α-2b (YPegIFN, Pegabin®, Tebao Pharmaceuticals Inc., China) in patients with CHC, we performed a comparative analysis of the early dynamics of HCVcAg and HCV RNA levels, and aimed to evaluate the early predictive efficacy of HCVcAg kinetics on SVR.
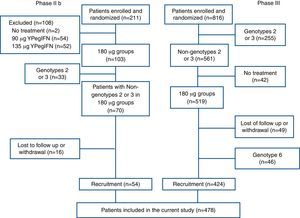
Patients and methodsPatientsAll patients were derived from phases IIb and III clinical trials of YPegIFN, which were multicenter, randomized, open-labeled, and positive drug-controlled (NCT01140997 and NCT01581398). According to the trials protocols all patients were aged 18 through 65 years, had positive anti-HCV and HCV RNA levels greater than 2000IU/mL at the last six months, and evidence of written informed consent. Exclusion criteria included significantly abnormal liver function, pregnancy or inability to practice adequate contraception, significant systemic or major illnesses other than liver disease, preexisting lower blood cells known history of antiviral or immunosuppressive therapy, and evidence of other viral infection.11 In addition, the current study included only patients infected with genotype 1 HCV and treated with 180μg PegIFN, and completed the planned treatment period.
A total of 211 and 816 patients were enrolled and randomized in the phases IIb and III clinical trials, respectively. In phase IIb trial, 157 patients were excluded due to use of other PegIFN, non-genotype 1 HCV infection, and loss-to-follow-up. In phase III trial, 392 patients were excluded due to non-genotype 1 HCV infection, no treatment initiation after randomization, loss-to-follow-up, and unavailability of HCVcAg data. At last, the cohort comprising 478 patients was analyzed in the present study (Fig. 1).
The study was approved by the ethical committees of Peking University People's Hospital and conducted according to the guidelines of the International Conference on Harmonization for Good Clinical Practices. The study protocol conformed to the ethical guidelines of the 1975 Helsinki Declaration.
HCV testsThe molecular HCV RNA assay is a confirmatory test for HCV infection using a reverse transcription-polymerase chain reaction (PCR). HCVcAg is detected in a two-step chemiluminescent microparticle immunoassay technology.13 The COBAS AmpliPrep/COBAS TaqMan automated real-time PCR platform (Roche Molecular Systems, Pleasanton, CA) was used to quantify HCV RNA levels. This assay has a LLOD of 15IU/mL and a lower limit of quantification of 43IU/mL. The Architect HCVcAg (Abbott Diagnostics, Wiesbaden, Germany) was used to quantify HCVcAg. As the manufacturer instructed, HCVcAg levels lower than 3.0fmol/L were considered non-reactive. HCV genotyping was based on hybridization of the amplified segment of the 5′ non-translated region of the HCV genome, and performed according to manufacturer's instructions (Abbott Diagnostic assay).
IFN therapy and definitions of response to IFN therapyAll patients were treated with YPegIFN or PegIFNα-2a (PegIFN) (180μg, once a week) in combination with RBV (<75kg, 1000mg daily and ≥75kg, 1200mg daily). The duration of treatment was 48 weeks for genotype 1 HCV infected patients and then followed up for 24 weeks.
SVR was defined as undetectable HCV RNA level (<15IU/mL) at week 24 after cessation of treatment, and rapid virologic response (RVR) was as undetectable HCV RNA at week 4 of therapy. Early virologic response (EVR) was defined as more than 2log10IU/mL decrease from baseline level after 12 weeks of therapy. Null response (NR) was considered when there was less than 2log10IU/mL decrease in HCV RNA level from baseline at week 12 of therapy. Reappearance of HCV RNA in serum after the end of therapy characterized relapse.14 N-SVR included null response and relapse. DHCVcAg and dHCVRNA were defined as a log10 reduction of serum HCVcAg and HCV RNA levels between other time points and baseline, respectively.
Statistical analysisQuantitative variables were expressed as mean±standard deviation or median (minimum, maximum), and categorical variables as absolute and relative frequencies. The categorical variables were compared between groups by the χ2 test or Fisher's exact test, and non-categorical variables were compared by Student's t-test, one-way ANOVA or the Mann–Whitney U-test. The optimal predictive values of HCVcAg and HCV RNA at different time points were assessed by calculating the area under the receiver operating characteristic (AUROC) curves. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated to determine the reliability of predictors of the response to therapy. All statistical analyses were performed with SPSS 16.0 (Chicago, IL, USA). Two-tailed p-values less than 0.05 were considered statistically significant.
All authors had access to the study data, and reviewed and approved the final manuscript.
ResultsA total of 478 patients were enrolled in the current study, of whom 475 (99.4%) patients were infected with genotype 1b HCV. There were 236 male and 242 female patients. The mean age was 41.52±12.23 years old. Mean baseline viral load and HCVcAg level were 6.00±0.79log10IU/mL and 3.42±0.74log10fmol/L, respectively. Baseline characteristics according to treatment outcome (SVR and N-SVR groups) are shown in Table 1. SVR patients had older age, higher baseline HCVcAg, and higher HCV RNA than N-SVR patients.
Baseline data for patients with sustained virological response (SVR) and non-sustained virological response (N-SVR).
| SVR | N-SVR | Statistics | p-Value | |
|---|---|---|---|---|
| Male/female (n) | 184/194 | 52/48 | χ2=0.349 | 0.555 |
| Age, mean±SD (years) | 41.52±12.00 | 46.45±12.34 | t=−3.631 | <0.001 |
| HCVcAg, median and range (log10fmol/L) | 3.66 (0.55, 4.30) | 3.78 (0.48, 4.30) | Z=−2.228 | 0.026 |
| HCV RNA, median and range (log10IU/mL) | 6.23 (3.58, 7.31) | 6.33 (3.34, 7.39) | Z=−2.379 | 0.017 |
| YPegIFN/PegIFNα-2a (n) | 241/137 | 63/37 | χ2=0.020 | 0.889 |
SVR, sustained virological response; N-SVR, non-sustained virological response; HCVcAg, hepatitis C virus core antigen; PegIFN, pegylated interferon; YPegIFN, type Y pegylated interferon α-2b.
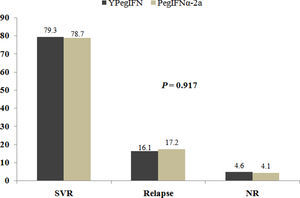
Of all participants in the current study, 304 and 174 were treated by 180μg YPegIFN plus RBV and PegIFNα-2a plus RBV, respectively. SVR was obtained in 79.3% of patients on YPegIFN, and in 78.7% in patients on PegIFNα-2a. Relapse occurred in 16.1% and 17.2% of patients receiving YPegIFN and PegIFNα-2a treatment, respectively (χ2=0.174, p=0.917, Supplementary figure).
Supplementary figure related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bjid.2015.04.007.
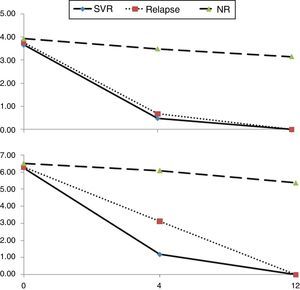
Early kinetics of HCVcAg and HCV RNA levels with different outcomesIn the first 12 weeks after initiation of antiviral therapy, kinetics of serum HCVcAg and HCV RNA were similar in SVR, relapsed and NR patients (Fig. 2). Baseline HCVcAg levels in SVR, relapsed, and NR patients were not significantly different, with median values of 3.66, 3.76 and 3.93log10fmol/L, respectively (χ2=2.933, p=0.231). The same was observed for baseline median HCV RNA (6.23, 6.28 and 6.51, χ2=1.953, p=0.377). Among the three groups of treatment response, the most rapid declines in serum HCVcAg and HCV RNA were observed in SVR patients, whereas NR patients showed the slowest decline.
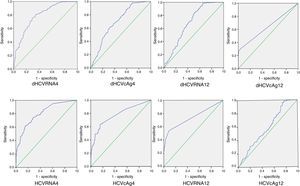
Accuracy of HCVcAg and HCV RNA declines at weeks 4 and 12 of treatment to predict SVRROC curves were used to determine the optimal cutoff values of HCVcAg and HCV RNA at weeks 4 and 12 that would maximize prediction of SVR (Fig. 3). Sensitivity, specificity, PPV and NPV were calculated using the optimal cutoff values (Table 2). Likewise, AUROCs of dHCVcAg, dHCVRNA, HCVcAg and HCV RNA were 0.695, 0.788, 0.775 and 0.803 at week 4, and 0.685, 0.688, 0.638 and 0.739 at week 12, respectively. As for dHCVcAg and HCVcAg, the highest AUROC for HCVcAg levels was obtained at week 4. At this time point the AUROC, based on 0.62 HCVcAg cutoff, was 0.775; the corresponding sensitivity, specificity, PPV and NPV were, 83.9%, 64.0%, 89.8%, and 51.2%, respectively. At week 4, HCVcAg and HCV RNA declines had similar PPVs (89.8% and 89.6%).
ROCs of values of HCVcAg and HCV RNA at weeks 4 and 12 to predict SVR. AUROCs were calculated to compare the values of HCVcAg and HCV RNA at weeks 4 and 12 to predict SVR and identify the best cutoff values. The random classifier line indicates a 50% post-test probability and the cutoff point represents the best compromise between sensitivity and specificity for the two assays.
Area under the ROC curve, sensitivity, specificity, and predictive values of sustained virological response based on total HCV core antigen and HCV RNA levels at weeks 4 and 12 after initial antiviral therapy.a
| dHCVcAg4 | dHCVRNA4 | HCVcAg4 | HCVRNA4 | dHCVcAg12 | dHCVRNA12 | HCVcAg12 | HCVRNA12 | |
|---|---|---|---|---|---|---|---|---|
| AUROC (95% CI) | 0.695 (0.632–0.758) | 0.788 (0.737–0.840) | 0.775 (0.718–0.832) | 0.803 (0.755–0.852) | 0.685 (0.618–0.752) | 0.688 (0.612 –0.755) | 0.638 (0.570 –0.707) | 0.739 (0.675–0.804) |
| Cutoff | 2.985 | 3.495 | 0.620 | 3.020 | 1.745 | 3.535 | 0.240 | 4.225 |
| Sensitivity | 0.593 | 0.809 | 0.839 | 0.831 | 0.958 | 0.984 | 0.955 | 1.000 |
| Specificity | 0.700 | 0.650 | 0.640 | 0.630 | 0.250 | 0.350 | 0.310 | 0.230 |
| PPV | 0.882 | 0.896 | 0.898 | 0.895 | 0.828 | 0.851 | 0.840 | 0.831 |
| NPV | 0.313 | 0.474 | 0.512 | 0.630 | 0.710 | 0.854 | 0.646 | 1.000 |
HCVcAg, hepatitis C virus core antigen; AUROC, area under the univariate receiver operating characteristic curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value
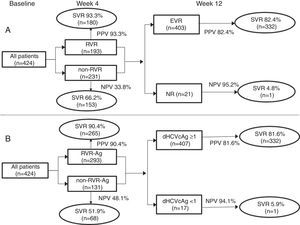
All 424 patients from the phase III trial were regarded as the model group. RVR-Ag had higher accuracy to predict SVR, with a PPV similar to RVR of HCV RNA (90.4% and 93.3%), and NPV of EVR-Ag was similar to EVR of HCV RNA (94.1% and 95.2%) (Fig. 4). Analysis on the validation group (54 patients from the phase IIb trial) demonstrated a PPV of 97.5% for RVR-Ag and a NPV of 100% for EVR-Ag.
Predictive efficacy of RVR and EVR of HCVcAg and HCV RNA at weeks 4 and 12. Analysis on the model group from the phase III clinical trial showed that undetectable HCVcAg at week 4 (RVR-Ag) had a high positive predictive value of SVR, and HCVcAg of more than 1log10IU/mL decrease at week 12 (EVR-Ag) had a high negative predictive value.
At week 4 time point, 333 (69.7%) out of 478 patients with negative HCVcAg (<3fmol/L), 137 out of 156 (87.8%) patients with <3fmol/L but detectable HCVcAg, and 163 out of 177 (92.1%) patients with undetectable HCVcAg achieved SVR. On the other hand, 448 out of 478 (93.7%) patients with negative HCVcAg (<3fmol/L) at week 12 time point, 14 out of 20 (70.0%) patients with <3fmol/L but detectable HCVcAg, and 361 out of 428 (84.3%) patients with undetectable HCVcAg achieved SVR. Although SVR rate was higher in patients with undetectable HCVcAg than in patients with negative but detectable HCVcAg, there was no difference between the two groups at either week 4 or week 12 (week 4: χ2=1.280, p=0.258; week 12: χ2=2.883, p=0.090).
DiscussionThe test for quantifying HCVcAg was developed in 1999 and has ever since been increasingly considered to reflect replication of HCV in different circumstances.15,16 However, despite being considered an alternative to HCV RNA assays, HCVcAg has not been recommended in clinical practice guidelines for diagnosis and guiding hepatitis C therapy.6,14 Its significance was unclear in clinical practice since it had not been tested in large clinical trials.
Based on phases IIb and III clinical trials on YPegIFN, 478 patients with genotype 1 HCV infection who completed the planned treatment duration were analyzed in the current study. All participants were treated with either 180μg YPegIFN or 180μg PegIFNα-2a both in combination with RBV. There were no significant differences in antiviral outcomes between YPegIFN and PegIFNα-2a groups. Therefore, we pooled the patients from both groups into one cohort. On the other hand, older age, higher baseline HCVcAg and HCV RNA levels were associated with SVR, which could not influence the comparative analysis of the accuracy of HCVcAg and HCV RNA kinetics to predict SVR.
At weeks 4 and 12 after initiation of antiviral therapy, similar kinetics patterns were observed for both serum HCVcAg and HCV RNA levels in CHC patients who had SVR, relapse or NR. Serum HCVcAg and HCV RNA declined most rapidly in the SVR group, and most slowly in the NR group suggesting that HCVcAg determination may be an alternative way to predict anti-HCV treatment outcomes. By calculating the AUROCs, we assessed the optimal SVR predictive cutoffs of HCVcAg and dHCVcAg, HCV RNA and dHCV RNA at weeks 4 and 12. We found that AUROCs of HCVcAg and dHCVcAg were smaller than those of HCVRNA and dHCVRNA at the same time points. As for HCVcAg, the biggest AUROC of 0.775 was seen for HCVcAg levels (not decline of HCVcAg) at week 4 for the 0.62 cutoff value. Such cutoff value yields PPV and NPV of 89.8% and 51.2%, respectively. At this time point, the PPV of HCVcAg was similar to that of HCV RNA decline (89.8% and 89.6%). On the other hand, RVR-Ag, which was defined as undetectable HCVcAg at week 4 of therapy, had higher accuracy for predicting SVR with a PPV of 90.4%. EVR-Ag, defined as more than 1log10IU/mL decline from baseline, had a PPV of 81.6% and a NPV of 94.1%. As for those patients with negative HCVcAg, undetectable HCVcAg was not significantly more predictive of SVR than negative but detectable HCVcAg.
In our previous study, we detected HCVcAg at 24, 48, 72, 96, 120 and 144h, and weeks 4, 8 and 12.11 We found that the highest AUROC and the best predictive time point was 144h after initiation of antiviral treatment, and the AUROCs decreased gradually at weeks 4, 8 and 12. Combining with the results of the present study in which the predictive accuracy of HCVcAg was comparable to that of HCV RNA at weeks 4 and 12 (that is RVR and EVR), we think that there are likely better predictive power and an earlier predictive time point for HCVcAg than for HCV RNA, but it needs further investigation. Alternatively, sensitive HCV RNA quantitative assays which meet guidelines for hepatitis C management, for example COBAS TaqMan HCV test and Abbott RealTime, are rare in developing countries including China, and due to its high cost and technical difficulties, HCV RNA testing is batched once or twice a week in many laboratories.17 Alternatively, we found that the predictive power of HCVcAg was superior to that of HCV RNA with LLOD of 500IU/mL which has been widely used in many areas of China because of its lower cost (data not shown). Furthermore, quantification of HCVcAg has some potential advantages as shown below. It can be performed with a simple operation and less time consuming (within 60min),18 does not require expert laboratory staff, and is less expensive than HCV RNA testing due to lower costs of equipment and reagents.15,18,19 HCVcAg is more stable and its detection is more reproducible than HCV RNA in clinical samples,9,20,21 and when performed in a fully automated chemiluminescent microparticle immunoassay has a very low intra- and inter-assay variability.22
Since 2011, triple therapies including the HCV NS3 protease inhibitors combined with PegIFN and RBV, in which response-guided therapy based on HCV RNA detection is applied, has been approved for those patients with chronic HCV genotype 1 infection.3,23 Moreover, IFN-free anti-HCV strategies are ongoing with promising results.24 Dual therapies of DAAs, like daclatasvir plus sofosbuvir, have been very successful with 98% SVR12 genotype 1 infected patients.25 In the era of DAAs, the clinical significance of HCVcAg needs to be investigated and its accuracy to predict treatment response will probably be adjusted.
In conclusion, serum HCVcAg could have the same early kinetics patterns as HCV RNA during treatment with PegIFN and ribavirin in CHC patients with SVR, relapse or NR. Although the sensitivity of HCVcAg testing was lower than that of HCV RNA, and AUROCs of HCVcAg and dHCVcAg were lower than those of HCV RNA and dHCV RNA, RVR and EVR of HCVcAg were comparable to those of HCV RNA, which means that HCVcAg may be as meaningful as HCV RNA in reflecting dynamic changes of viral replication and in predicting SVR earlier during double treatment. As a reliable, faster, more cost-effective, and less labor-intensive alternative to HCV RNA testing, HCVcAg could be a promising assay in guiding antiviral treatment, especially in low- and middle-income countries.
Conflicts of interestLW has received grant/research support from Roche, Novartis, Bristol-Myers Squibb and Abbott. The other authors declared no conflict of interest.
This study was supported by National S&T Major Project for Infectious Diseases Control (grant no. 2012ZX10002-003), National Major S&T Special Project for “Significant New Drugs Development” (grant no. 2012ZX09303-019), and Beijing Natural Science Foundation (grant no. 7122191). We would like to acknowledge the assistance of Abbott Diagnostics in the provision of the HCV core antigen kits for this evaluation.