Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most frequently isolated agents in both nosocomial and community settings. It is a constant challenge for antibacterial therapy. Therefore, it becomes essential to understand the epidemiology of MRSA isolates in the institution and/or region to guide empirical therapy. The objective of this study was to evaluate the epidemiological characteristics of MRSA isolates in the state of Santa Catarina, Brazil, and determine if there is a clonal spread. We evaluated 124 clinical isolates of MRSA obtained from various anatomical sites from patients in the state of Santa Catarina in Southern Brazil. The antimicrobial susceptibility profile was evaluated by disk diffusion and minimum inhibitory concentration (MIC) was determined by Etest and broth macrodilution. SCCmec types were determined by multiplex PCR and the clonal relationship among isolates was assessed by pulsed field gel electrophoresis. Antimicrobials that have demonstrated lower rates of resistance were tetracycline (20.2%), sulfamethoxazole–trimethoprim (20.2%) and chloramphenicol (12.9%). We did not detect any resistance to glycopeptides, daptomycin, linezolid, and tigecycline. SCCmec type III was predominant (54%), followed by type II (21.8%), consistent with other Brazilian studies. Twenty-six clones were observed grouping 72 (58%) isolates and no clonal relationship was observed between our isolates and the major epidemic clones circulating in Brazil. An intriguing distinct MRSA epidemiology was observed in Santa Catarina, compared to other Brazilian regions.
Staphylococcus aureus is a major agent of community-acquired infections and healthcare associated infections. A multicenter study among Brazilian hospitals during 2005–2008 found that S. aureus was the main agent of bloodstream infections (20.2%) and infections of the skin and soft tissue (28.1%) and the second most common agent in pneumonia of hospitalized patients (24.9%). Of these infections, approximately 30% were resistant to methicillin (MRSA).1
Staphylococci become resistant to methicillin by changing transpeptidases in the wall, called penicillin-binding proteins (PBPs), and lose affinity for all β-lactam agents. This information is contained in the mecA gene responsible for encoding an altered PBP (PBP2a).2,3
Until the early 1990s, MRSA isolates were restricted to hospitals. Currently, however, MRSA is no longer exclusively associated with healthcare infections. During this period, increase in community-acquired MRSA (CA-MRSA) among patients without assignable risk factors for acquiring MRSA was observed (i.e., had no direct or indirect contact with health services that could associate the MRSA infection with health care) .4
Since then, a reverse phenomenon began to occur: community isolates, once characterized by the presence of a staphylococcal cassette chromosome (SCCmec) type IV, began to be isolated in hospital settings and those types that were typically isolated in the hospital began to appear in outpatients.5,6
As the epidemiology of MRSA proved to be dynamic, it is important to generate data to better understand the evolution of this epidemiological behavior and to assess its impact on clinical settings.7 Santa Catarina State has an intriguing low prevalence of MRSA compared to other Brazilian States and understand this epidemiological dynamics would be valuable.
The objective of this study was to evaluate the epidemiological characteristics of MRSA isolates in the state of Santa Catarina.
MethodsBacterial samplesWe used 124 clinical MRSA isolates obtained from various anatomical sites from patients attended in two cities (Blumenau and Florianopolis), located in the State of Santa Catarina, Southern Brazil. Samples were collected from November 2009 through February 2013. One isolate per patient was considered. Samples were selected by convenience. Identification was done using standardized phenotypic methodology8 and isolates were maintained frozen (−20°C) in skim milk (DIFCO®) plus 10% glycerol.
Antimicrobial susceptibility testingAntimicrobial susceptibility testing was performed using the disk diffusion method, according to the recommendations and interpretative criteria of the Clinical and Laboratory Standards Institute9 and European Committee on Antimicrobial Susceptibility Testing (to tygecycline only).10 The antimicrobials tested were gentamicin (10μg), ciprofloxacin (5μg), erythromycin (15μg), clindamycin (2μg), trimethoprim/sulfamethoxazole (1.25μg/23.75μg), chloramphenicol (30μg), tetracycline (30μg), teicoplanin (30μg), tigecycline (10μg), and linezolid (30μg). MRSA was characterized using the cefoxitin disk (30μg), and confirmed by PCR for the gene mecA.11 To detect β-lactamase production, we used nitrocefin disk (BD BBL™ DrySlide™ Nitrocefin), using bacterial suspensions in physiological saline. S. aureus ATCC 29213 was used as a positive control, and S. aureus ATCC 25923 was used as a negative control.
MIC determinationVancomycin MICs were determined by macrodilution method9 and by Etest® (BioMérieux, Marcy l’Etoile, France) following CLSI interpretative criteria9 and the manufacturer's instructions, respectively. MICs for teicoplanin and daptomycin were determined by Etest®. S. aureus strains ATCC 29213 (MSSA), ATCC 43300 (MRSA), ATCC 700698 (hVISA) and ATCC 700699 (VISA) were used for quality control.
Multiplex PCRThe SCCmec type was determined using the multiplex PCR method according to the protocol developed by Zhang et al.12 The amplicons that were formed had the following sizes: I (613bp), II (398bp), III (280bp), IVa (776bp), IVb (493bp), IVc (200bp), IVd (881bp), and V (325bp).12,13
For isolates presenting phenotypic inducible resistance to clindamycin, erm gene was assessed by a multiplex PCR protocol, described elsewhere.14 The PCR product (610bp for ermA and 520bp for ermC) was analyzed by electrophoresis through a 1.5% agarose gel.14,15
Pulsed-field gel electrophoresis (PFGE)PFGE was performed according to McDougal et al.16 and Pinto et al.17 The fragments were subjected to PFGE using 1% agarose gels (Pulsed Field Certified Agarose; Bio-Rad) in 0.5× Tris–borate–EDTA buffer with a CHEF-DR III system (Bio-Rad). Gels were stained with 0.5μg/mL ethidium bromide, visualized under UV light, and photographed using a GelDoc™ XR System (Bio Rad). PFGE patterns were analyzed using Bionumerics version 6.1 (Applied Maths, Sint-Martens-Latem, Belgium) and clustered by UPGMA. A dendrogram was generated from a similarity matrix calculated using the Dice similarity coefficient with an optimization of 0.5% and a tolerance of 1%. PFGE clusters were defined as isolates with a similarity of 80% or higher on the dendrogram.18 SCCmec control strains were included [type I (NCTC10442); type II (N315); type III (85/2082); type IVa (CA05); type IVb (8/6-3P); type IVc (MR108); type IVd (JSC4469) and type V (WIS)], as well as isolates of the main circulating epidemic clones in Latin America (Brazilian Epidemic Clone, New York/Japan, USA300, USA400, Pediatric, OSPC, Cordobean, E-MRSA 15, E-MRSA 16).
ResultsMRSA isolates were recovered mainly from lower respiratory tract samples (37.1%), followed by clinical osteomyelitis cases (19.4%), blood (13.7%), surgical wounds (9.7%), urine (4%), and abscesses (3.2%). In addition, 1.6% of MRSA were from surveillance cultures, and 11.3% were recovered from other anatomical sites. Considering the origin of the samples, 17 (13.7%) were considered community isolates and 107 (86.3%) hospital isolates.
MRSA presented the following resistance rates: amikacin (35.5%), gentamicin (33.1%), chloramphenicol (12.9%), tetracycline (20.2%), ciprofloxacin (79%), norfloxacin (72.6%), sulfamethoxazole–trimethoprim (20.2%), clindamycin (75%), and erythromycin (81.5%). Also, 93 strains (75%) showed production of β-lactamase. All isolates were susceptible to linezolid and tigecycline.
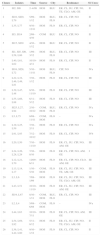
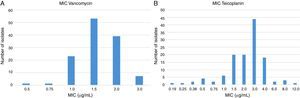
All isolates were susceptible to vancomycin and MICs are presented in Fig. 1. Seven isolates showed MIC of 3.0μg/mL by Etest. By macrodilution three of them showed MIC of 1.0μg/mL and four had MIC of 2.0μg/mL. Etest also overestimated (above CLSI susceptibility breakpoint) teicoplanin MIC for one isolate but macrodilution defined its MIC as 4μg/mL (susceptible by CLSI interpretative criteria).
MIC of the glycopeptides to 124 MRSA determined by Etest. In (A), the values obtained for vancomycin. The seven isolates with MIC of 3.0μg/mL were considered susceptible because all MICs were ≤2.0μg/mL by microdilution. In (B), the results for teicoplanin. One isolate (MIC 12μg/mL) when tested by microdilution yielded an MIC of 4.0μg/mL (susceptible).
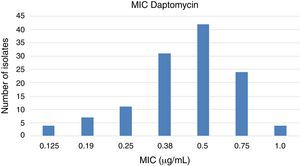
Daptomycin, an alternative to glycopeptides for the treatment of infections caused by MRSA, showed MCI values within the susceptibility range (Fig. 2), even though four isolates presented MIC of 1.0μg/mL (upper limit of susceptibility category).
Although predominant SCCmec was type III (54%, 67/124), there was a heterogeneous distribution of SCCmec types: type II (21.8%), type IVa (10.5%), type IVc (3.2%) and type IVb (2.4%). Five isolates carried types I and II together. Among CA-MRSA (n=17), only four showed SCCmec type IV: two IVa and two IVb. Among the other 13 isolates, eight had SCCmec type III and five had type II.
Of the 124 isolates, 23 (18.5%) had a positive D test: 20 (87%) had ermA gene and three (13%) carried ermC. Twelve (9.7%) isolates presented resistance to erythromycin and susceptibility to clindamycin (negative D test) and had no positive PCR reaction. Fifty-six (45.1%) isolates have had constitutive resistance mechanism and only 33 (26.6%) were susceptible to both antibiotics.
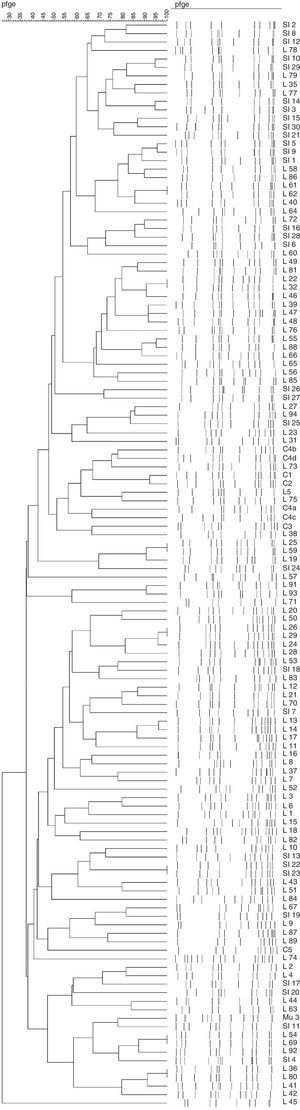
Twenty-six clones (Fig. 3) were observed, grouping 72 isolates (with five being the highest number of isolates per clone). Indeed, 52 isolates presented a unique pulsotype. Isolates belonging to all clones carried SCCmec type III (Table 1).
Epidemiological and antibiotic susceptibility characteristics of the 26 clones.
| Clones | Isolates | Time | Source | City | Resistance | SCCmec |
|---|---|---|---|---|---|---|
| 1 | SI2, SI8 | 2–4/09 | HOS | BLU | ER, CL, SU, CIP, N0, TE, CLO, AM, GE | II |
| 2 | SI10, SI29, L79 | 5/09–1/12 | HOS | BLU, FLO | ER, CL, CIP, NO | II |
| 3 | L35, L77 | 9/10–12/11 | HOS | FLO | ER, CL, CIP, NO | II |
| 4 | SI3, SI14 | 2/09–4/10 | COM | BLU | ER, CL, CIP, NO | II |
| 5 | SI15, SI30 | 4/10–1/12 | HOS | BLU | ER, CL, CIP, NO | II |
| 6 | SI1, SI5, SI9, L58, L86 | 1/09–3/12 | HOS | BLU, FLO | ER, CL, CIP, NO | III |
| 7 | L40, L61, L62 | 10/10–4/11 | HOS | FLO | ER, CL, CIP, NO | II |
| 8 | SI16, SI28, L72 | 5/10–11/11 | HOS | BLU, FLO | CIP, NO | IVa |
| 9 | L22, L32, L46, L49, L81 | 7/10–1/12 | HOS | FLO | ER, CL, CIP, NO | III |
| 10 | L39, L47, L48 | 8/10–11/10 | HOS | FLO | ER, CL, CIP, NO | III |
| 11 | L55, L66, L88 | 6/11–5/12 | HOS | FLO | ER, CL, CIP, NO | III |
| 12 | SI25, L27, L94 | 2/10–10/12 | COM, HOS | BLU, FLO | ER, CL, CIP, NO | IVa |
| 13 | L5, L75 | 8/08–11/11 | COM, HOS | FLO | – | IVa |
| 14 | L19, L25, L59 | 7/10–2/11 | HOS | FLO | ER, CL, CIP, NO | IVa |
| 15 | L91, L93 | 7/12–10/12 | HOS | FLO | ER, CL, CIP, NO | IVb |
| 16 | L20, L50 | 7/10–11/10 | HOS | FLO | ER, CL, SU, CIP, NO, AM. GE | III |
| 17 | L24, L26, L28, L29 | 7/10–8/10 | HOS | FLO | ER, CL, CIP, NO, AM, GE | I |
| 18 | L12, L21, L70 | 12/09–6/11 | HOS | FLO | ER, CL, CIP, NO, CLO, AM, GE | III |
| 19 | L13, L14, L17 | 1/10–5/10 | COM, HOS | FLO | ER, CL, SU, CIP, NO, TE, AM, GE | III |
| 20 | L3, L6 | 7/08–1/09 | HOS | FLO | ER, CL, SU, CIP, NO, TE, CLO, AM, GE | III |
| 21 | L43, L51 | 10/10–11/10 | HOS | FLO | ER, CL, SU, CIP, NO. AM. GE | III |
| 22 | SI19, L67 | 8/10–1/11 | COM, HOS | BLU, FLO | ER, CL, CIP, NO | III |
| 23 | L2, L4 | 5/08–6/08 | COM, HOS | FLO | – | IVa |
| 24 | L44, L63 | 10/10–7/11 | HOS | FLO | ER, CL, CIP, NO, AM, GE | III |
| 25 | L54, L69, L92 | 5/11–8/12 | HOS | FLO | ER, CL, SU, CIP, NO, TE, CLO, AM, GE | II |
| 26 | L36, L41, L42, L80 | 9/10–1/12 | HOS | FLO | ER, CL, CIP, NO | II |
HOS, hospital; COM, community; BLU, blumenau; FLO, florianopolis; ER, erythromycin; CL, clindamycin; SU, trimethoprim/sulfamethoxazole; CIP, ciprofloxacin; NO, norfloxacin; TE; tetracycline; CLO, chloramphenicol; AM; amikacin; GE, gentamycin.
The prevalence of MRSA in the state of Santa Catarina is extremely low and is comparable to rates found in Scandinavian countries.2,19,20 Isolation rates are in the range of 4–8% for all isolated S. aureus and these rates are even lower when only nosocomial isolates are considered (less than 2%). Such low occurrence of MRSA justifies the intriguing low number of MRSA included in this study, as the two most populous cities of Santa Catarina were included. In a report published by the World Health Organization (WHO), several European countries have national surveillance data with very low rates of MRSA: Denmark (1.2%), Estonia (1.7%), Finland (2.8%), Iceland (2.8%), Netherlands (1.4%), Norway (0.3%) and Sweden (0.8%).21 These countries may have succeeded in maintaining low MRSA rates because of effective search-and-destroy policies and/or controlled antibiotic overuse.22 Of note, occurrence of MRSA in Santa Catarina differs greatly from other regions of Brazil, which have, in general, MRSA rates of around 29%.23,24 As far as we know, those search-and-destroy policies are not practiced in Santa Catarina and, surprisingly, MRSA isolation rates are similar to those found in the Scandinavian countries.
There was a predominance of isolates from lower respiratory tract infections, osteomyelitis, and bloodstream infections. These data are quite distinct from those found in studies with similar characteristics, which demonstrate a predominance of bloodstream infections (39%)25 or skin and soft tissue infections (61.5%).26
Compared to the antimicrobial surveillance in the multicenter study SENTRY,1 resistance rates in this study were much lower: ciprofloxacin (91.4–79%), tetracycline (46.7–20.2%), trimethoprim–sulfamethoxazole (68.1–20.2%), clindamycin (87.9–75%) and erythromycin (94–81.5%). A possible explanation for the large differences in rates of susceptibility could be due to the epidemiological profile. Cavalcante et al. proposed phenotypic markers associated with SCCmec types. Resistance to tetracycline and trimethoprim–sulfamethoxazole may be associated with SCCmec type III, in which 100% resistance was found to both drugs. SCCmec type IV had absolute susceptibility, with only 2% of the isolates resistant to trimethoprim–sulfamethoxazole and 100% susceptibility to tetracycline.27 We found a predominance of SCCmec type III, which justifies why the resistance rates in our study were lower than those found in the rest of Brazil.
The presence of β-lactamase may result in unusual phenotypes known as BORSA (Borderline Oxacillin Resistant S. aureus). These bacteria, despite lacking the mecA gene, may be resistant to oxacillin and are phenotypically characterized as MRSA.28 They may also be responsible for elevated MICs of oxacillin. After therapy with β-lactams, these bacteria may induce the development of vancomycin resistance, a phenotype called β-lactam antibiotics-induced vancomycin resistant S. aureus (BIVR).29
All MRSA were susceptible to vancomycin, teicoplanin and daptomycin. Although the phenotype of resistance to these drugs is uncommon.30 31.45% of the isolates showed MIC of 2.0μg/mL for vancomycin, which may suggest the development of a phenomenon of an upward trend for MICs for vancomycin, called “MIC creep”.31
The isolates demonstrated higher prevalence of gene ermA than ermC. The D test proved to be a methodology capable of detecting all isolates with inducible clindamycin resistance. Therefore, the test is easy to perform, inexpensive and is clinically relevant because it reduces the risk of inappropriate antibiotics use.32
In our study, there was a high prevalence of SCCmec type III (54%). In 2010, a study conducted at the Hospital de Clinicas de Porto Alegre (capital of the neighboring state of Rio Grande do Sul), a high prevalence of SCCmec type III was detected (49%),33 similar to our findings. In 2005, in contrast to the local epidemiology, a study conducted at the Hospital de Clinicas de São Paulo found that 65% of MRSA carried SCCmec type IV.34
Not surprising, our isolates did not present a clonal dissemination, as both cities (Florianopolis and Blumenau) are separated by approximately 120km and the isolates were collected during a period of three years.
Interestingly, we did not find clonal relationship between our isolates and the major circulating epidemic clones in Brazil, which may suggest that the state of Santa Catarina has different epidemiological characteristics compared to other regions in the country. Surveillance studies must be continuously conducted to better understand epidemiological dynamics of such a clinically relevant microorganism.
ConclusionsAlthough we identified the SCCmec that were present and evaluated the presence of clones, the present study did not provide consistent scientific evidence for the low prevalence of MRSA in Santa Catarina, which was very different from the data found in other Brazilian states and some European countries.
We conclude that the MRSA epidemiological profile in Santa Catarina is similar to that found in the rest of Brazil, with a predominance of SCCmec type III, but the resistance rates were lower than those found in the rest of Brazil.
A wide variety of complexes with clonal isolates were detected without evidence of clonal spread, and a small amount of high genetic diversity was found.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Pesquisa e Desenvolvimento Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Rio Grande do Sul (FAPERGS) for financial support. Thanks to Laboratório Santa Luzia (Cassia Zoccoli and Nina Tobouti) and Laboratório Santa Isabel (Reginaldo Simões and Marcelo Molinari) for granting clinical samples.