In Brazil, bats have been assigned an increasing importance in public health as they are important rabies reservoirs. Phylogenetic studies have shown that rabies virus (RABV) strains from frugivorous bats Artibeus spp. are closely associated to those from the vampire bat Desmodus rotundus, but little is known about the molecular diversity of RABV in Artibeus spp. The N and G genes of RABV isolated from Artibeus spp. and cattle infected by D. rotundus were sequenced, and phylogenetic trees were constructed. The N gene nucleotides tree showed three clusters: one for D. rotundus and two for Artibeus spp. Regarding putative N amino acid-trees, two clusters were formed, one for D. rotundus and another for Artibeus spp. RABV G gene phylogeny supported the distinction between D. rotundus and Artibeus spp. strains. These results show the intricate host relationship of RABV's evolutionary history, and are invaluable for the determination of RABV infection sources.
Rabies is a worldwide neglected fatal encephalitis,1,2 which is listed amongst the ten major infectious causes of human deaths worldwide, estimated at 55,000 per year.3 This infection is efficiently preventable by vaccination,4 but treatment costs of rabid or exposed patients, diagnostic procedures, and vaccines make it a significant challenge for public health systems in endemic countries.5,6
The disease is caused by rabies virus (RABV) (family Rhabdoviridae, genus Lyssavirus),7 an enveloped virus with a length of 110-250nm and a diameter of 75nm, usually transmitted by the saliva of an infected mammal. The RABV genome has a negative-stranded non-segmented ssRNA with 11,932-nucleotides that encodes the five structural proteins N (nucleoprotein), P (phosphoprotein), M (matrix), G (envelope glycoprotein), and L (large, RNA-dependent RNA-polymerase).8,9Between 2004 and 2005, 62 people died in the Brazilian Amazon of rabies transmitted by Desmodus rotundus vampire bat, a primary reservoir of rabies in Latin America.10,11 Regarding frugivorous bats of the genus Artibeus, rabies has been reported in the species A. fimbriatus, A. jamaicensis, A. lituratus, and A. planirostris.12Artibeus spp. bats have been assigned an increasing importance in public health, as they are considered a rabies reservoir for humans in urban areas in Brazil, which is aggravated by the increasing population of these bats and the fact that they share roosts with D. rotundus.12–14
From 2003 to 2008, the Instituto Pasteur in Brazil tested 18,007 non-hematophagous bats for rabies, 252 of which were found to be positive. Also, from 2005 and 2007, 56 out of 160 non-hematophagous bats that tested positive for rabies were classified as A. lituratus.
Phylogenetic studies based on the N gene have suggested that RABV lineages from Artibeus sp. are not divergent from those from D. rotundus,15 all belonging to the same genic16 and antigenic variant 3 (AgV3).17 Nonetheless, these studies have provided only inconclusive results, as they were based on a very restricted sampling regarding geographic area and sample number, mainly in the case of Artibeus spp. In addition, these studies were based on a single gene sequences for phylogenetic reconstructions.
The ability to determine the source of infection and the epidemiology of rabies cycles are paramount for accurate decision-making in public health, mainly regarding vaccination strategies and animal population control. This study aimed to evaluate the possibility of distinction between RABV genetic lineages related to D. rotundus and Artibeus spp. bats based on N and G genes sequences.
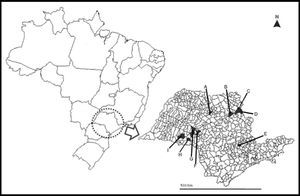
Materials and methodsRabies virus strainsTwenty Artibeus spp. RABV strains were obtained from first-passage isolates in mice inoculated with 20% suspensions of A. lituratus and Artibeus spp. central nervous systems (CNS); 15 D. rotundus-related strains were obtained directly from naturally infected cattle brain tissues (Table 1). These 35 strains were collected in nine municipalities from São Paulo State, Southeastern Brazil (Fig. 1), between 2004 and 2005. All samples were diagnosed positive for rabies by direct immunofluorescence test (DIFT) targeted to the viral nucleoprotein.18
GenBank accession numbers for the reference sequences of N gene and G gene used in phylogenetic analysis in this study showing strain, the specific-host from which the AgV3 RABV lineages were isolated, city, and year in which the samples were obtained.
| GenBank number | Strain | City | Specific-host | Year | |
|---|---|---|---|---|---|
| N gene | G gene | ||||
| JF682392 | JF682427 | 05/Art3250 | Catanduva | Artibeus spp. | 2005 |
| JF682393 | JF682428 | 05/Art4578 | Monte Mor | Artibeus lituratus | 2005 |
| JF682394 | JF682429 | 05/Art3598 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682395 | JF682430 | 05/Art3738 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682396 | JF682431 | 05/Art4850 | Ribeirão Preto | Artibeus spp. | 2005 |
| JF682397 | JF682432 | 05/Art4932 | Ribeirão Preto | Artibeus spp. | 2005 |
| JF682398 | JF682433 | 05/Art5459 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682399 | JF682434 | 05/Art6734 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682400 | JF682435 | 05/Art6956 | Ribeirão Preto | Artibeus spp. | 2005 |
| JF682401 | JF682436 | 05/Art7045 | Ribeirão Preto | Artibeus spp. | 2005 |
| JF682402 | JF682437 | 05/Art7436 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682403 | JF682438 | 05/Art8688 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682404 | JF682439 | 05/Art8921 | Ribeirão Preto | Artibeus lituratus | 2005 |
| JF682405 | JF682440 | 05/Art10509 | Ribeirão Preto | Artibeus spp. | 2005 |
| JF682406 | JF682441 | 05/Art7270 | Paraguaçú Paulista | Artibeus lituratus | 2005 |
| JF682407 | JF682442 | 05/Art7547 | Paraguaçú Paulista | Artibeus lituratus | 2005 |
| JF682408 | JF682443 | 05/Art8456 | Paraguaçú Paulista | Artibeus lituratus | 2005 |
| JF682409 | JF682444 | 05/Art11206 | Paraguaçú Paulista | Artibeus lituratus | 2005 |
| JF682410 | JF682446 | 05/Art8639 | Marília | Artibeus lituratus | 2005 |
| JF682413 | JF682445 | 05/Art7848 | Marília | Artibeus spp. | 2005 |
| JF682411 | JF682460 | 05/Bov6314 | Garça | Bovine | 2005 |
| JF682412 | JF682461 | 05/Bov7535 | Platina | Bovine | 2005 |
| JF682414 | JF682447 | 05/Bov451 | Altinópolis | Bovine | 2005 |
| JF682415 | JF682449 | 05/Bov3924 | Altinópolis | Bovine | 2005 |
| JF682416 | JF682451 | 05/Bov10339 | Altinópolis | Bovine | 2005 |
| JF682417 | JF682450 | 04/Bov8967 | Altinópolis | Bovine | 2004 |
| JF682418 | JF682448 | 04/Bov2196 | Altinópolis | Bovine | 2004 |
| JF682419 | JF682452 | 05/Bov2579 | Santo Antônio da Alegria | Bovine | 2005 |
| JF682420 | JF682453 | 04/Bov3441 | Santo Antônio da Alegria | Bovine | 2004 |
| JF682421 | JF682454 | 04/Bov3833 | Santo Antônio da Alegria | Bovine | 2004 |
| JF682422 | JF682455 | 04/Bov4698 | Santo Antônio da Alegria | Bovine | 2004 |
| JF682423 | JF682456 | 04/Bov6930 | Santo Antônio da Alegria | Bovine | 2004 |
| JF682424 | JF682457 | 04/Bov11044 | Santo Antônio da Alegria | Bovine | 2004 |
| JF682425 | JF682458 | 04/Bov11817 | Santo Antônio da Alegria | Bovine | 2004 |
| JF682426 | JF682459 | 04/Bov11818 | Santo Antônio da Alegria | Bovine | 2004 |
A map of São Paulo State (minor map) and Brazil (larger map), showing the cities (in black) were the 35 bats used in this study were collected. A) Catanduva, B) Ribeirão Preto, C) Altinópolis, D) Santo Antônio da Alegria, E) Monte Mor, F) Garça, G) Marília, H) Platina, and I) Paraguaçú Paulista.
The nucleoprotein and the glycoprotein sequences generated in this study have been assigned the GenBank accession numbers JF682392 - JF682426 and JF682427-JF682461, respectively.
Amplification and sequencing of the nucleoprotein and glycoprotein genesTotal RNA from the 35 RABV isolates CNS samples tested and the from positive and negative controls were extracted with TRIzol™ (Invitrogen – Carlsbad, CA, USA) method, following the manufacturer's instructions. The challenge virus standard (CVS) fixed strain of RABV isolated in mice brain and nuclease free-water were used as positive and negative controls, respectively.
Reverse transcription polymerase chain reaction (RT-PCR) to partially amplify the N and G genes was performed according to the protocol described by Carnieli et al.,19 using the primers described by Orciari et al.20 for the N gene and those described by Sato et al.21 for the G gene (Table 2). The PCR products were purified from the PCR reactions using the QIAquick™ Gel Extraction KitTM (Qiagen – Valencia, CA, USA), according to the manufacturer's instructions. The products with nonspecific bands were purified using 1% agarose gel and the QIAquick® kit. After the purification step, the DNA samples were visually quantified in 2% agarose gel with Low Mass DNA Ladder (Invitrogen – Carlsbad, CA, USA), following the manufacturer's instructions.
Primers for RT-PCR and genetic sequencing of RABV N and G genes.
| Primers | Orientation | Sequence | Gene |
|---|---|---|---|
| 21G | Sense | 5’ ATGTAACACCTCTACAATG 3’ | N |
| 304 | Antisense | 5’ TTDACGAAGATCTTGCTCAT 3’ | N |
| GA 3222-40 | Sense | 5¿CGCTGCATTTTRTCARAGT 3¿ | G |
| GB 4119-39 | Antisense | 5’ GGAGGGCACCATTTGGTMTC 3’ | G |
| GS 3994 | Sense | 5’GGGMTTTGTGGATGAAAGRGGC 3’ | G |
| GantiBR2072 | Antisense | 5’ TGCTGATTGCRCCTACATT 3’ | G |
The DNA sequencing reaction mixture consisted of 4μL of BigDye 3.1™ (Applied Biosystems – Foster City, CA, USA), 3.2pmol of both sense and antisense primer for each gene in separate reactions, 30–60ng of target DNA and DNase-free water to a final reaction of 10μL. The reaction was performed in a Mastercycler Gradient thermal cycler™ (Eppendorf, NY, USA) with 35 cycles at 96°C for 10s, 50° C for 5s, and 60°C for 4min, with a ramp of 1°C/s between each temperature. Sequencing reaction products were purified with Sephadex™G-50 fine beads (GE Healthcare Biosciences) in 96-well multiscreen HV plates. After purification, the sequences were resolved in an ABI-3130™ Automatic Sequencer (Applied Biosystems – Foster City, CA, USA).
Phylogenetic analysesTo construct the genealogic trees, nucleotide and putative amino acids sequences for the 35 RABV strains of N and G genes were aligned by the CLUSTAL/W multiple alignment algorithm method using the BioEdit program,22 and then by manually checking the alignments for each set of aligned sequences. A score was assigned to each of the nucleotides shown on the electropherograms for each of the sequencing reactions using the online Phred application. Only positions with a Phred score >20 were used.23 The final sequence for each strain was obtained using the Contig Assembly Program (CAP) in BioEdit v.5.0.922 and submitted to BLASTn for homology confirmation.
Phylogenetic trees of the RABV isolates were built using the Neighbor-joining algorithm and the maximum composite likelihood (MCL) evolutionary model implemented in Mega 4.1 (© 1993–2008)24 with 1,000 bootstrap replicates. Additionally, 38 homologous sequences recovered from GenBank were included in the phylogenetic trees of the N and G genes (29 and none, respectively) and European bat Lyssavirus 1 (another specie in the Lyssavirus) was used as an outgroup.
The minimum, maximum, and mean nucleotide (with the MCL model) and amino acids (with the Poisson correction) identities for the clusters for the N and G gene sequences were calculated using Excel (©1985–2003 Microsoft Corporation) based on the identity matrices calculated with the BioEdit program. The changes in the amino acids observed in the samples analyzed were studied using Mega 4.1 (© 1993–2008)24 and BioEdit v.7.0.022 programs.
ResultsAfter editing, the N gene was 1,281-nucleotides long, located between nucleotides 68 and 1,350 in relation to the N gene CVS reference strain (GenBank number AF406696) and had a putative amino acid sequence with 427 amino acids. Regarding the G gene, sequences were 1,571-nucleotides long, located between nucleotides 8 and 1,579 in relation to the G gene CVS reference strain (GenBank number FJ979833) and had a putative amino acid sequence with 520 amino acids.
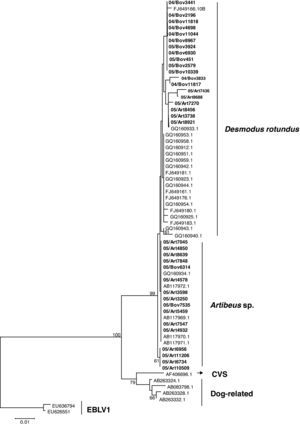
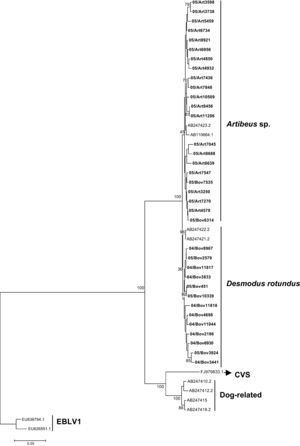
The N gene nucleotide phylogenetic tree showed three clusters for the 35 RABV strains included in the study; one cluster for D. rotundus strains and two for the Artibeus spp. strains. Regarding N protein amino acids tree, the 35 strains segregated in only two clusters, one for D. rotundus and one formed mainly by Artibeus spp. strains (Fig. 2). Nucleotides and amino acids identities for the N region under analysis between D. rotundus and Artibeus spp. sequences ranged from 97.4% to 98.7% and 98.1% to 99.7%, respectively.
RABV N amino acids tree showing clusters specific to Desmodus rotundus and Artibeus spp. bats. The tree was built with the Neighbor-joining distance algorithm and the Poisson correction with 1,000 bootstrap replicates and European bat Lyssavirus (EBLV-1) as outgroup. The bar represents the number of substitutions per site.
The G gene nucleotide phylogenetic tree showed two clusters for the 35 RABV strains included in the study; one cluster for D. rotundus strains and one for the Artibeus spp. strains. Regarding G amino acids tree, the 35 strains segregated in two clusters, one for D. rotundus and one formed by Artibeus spp. strains (Fig. 3). Nucleotides and amino acids identities for the G region under analysis between D. rotundus and Artibeus spp. sequences ranged from 97.0% to 99.1% and 96.1% to 99.0%, respectively.
RABV G amino acids tree showing clusters specific to Desmodus rotundus and Artibeus spp. bats. The tree was built with the Neighbor-joining distance algorithm and the Poisson correction with 1,000 bootstrap replicates and European bat Lyssavirus (EBLV-1) as outgroup. The bar represents the number of substitutions per site.
Frugivorous bats of Artibeus spp. have an emerging importance for rabies epidemiology in Brazil, mainly in urban centers, and have been reported as carriers of RABV lineages close to those found in the vampire bat D. rotundus.12,16 In the present investigation, RABV N and G phylogenies of strains recovered from these bats showed the existence of viral lineages that can be accurately attributed to D. rotundus or Artibeus spp. bats, a previously unknown fact.
RABV lineages heterogeneity expressed phylogenetically as host-specific lineages is a widely documented epidemiological phenomenon,16,19,25,26 and is influenced by geographic barriers rather than by species barriers only.27 Accordingly, the two Artibeus spp. clusters found for N gene sequences might also represent regional sub lineages of RABV, a fact already described for D. rotundus RABV lineages in Brazil.28,29 These observations agree with the proposition that, regarding rabies in bats, host species are as important as geographic variations.30 Regarding viral lineages, genetic variations that occur within a host species are different from those that occur in another, and this variation, coupled with the host's geographical isolation, may explain the RABV genetic differences reported in this work.31
Seven strains (05/Art7270, 05/Art8456, 05/Art7436, 05/Art8921, 05/Art8688, 05/Art3738, and 08IacriSP3577B) were classified in the Artibeus spp. group for the N nucleotides tree; however they segregated into the D. rotundus cluster of the N amino acids tree. This fact suggests the possibility that Artibeus spp. strains are still under an adaptation process after the spill-over event from D. rotundus, as already suggested by Kissi,31 who have already experimentally reported this type of stepped adaptation of RABV. This phenomenon can occur as a consequence of the predominance of synonymous mutations over non-synonymous nucleotides mutations, which leads to greater differences among the nucleotide sequences than among the amino acids sequences.32,33
Regarding the Bov7525 and Bov6314 strains, in cattle, both segregated in the Artibeus spp. RABV cluster; this was unexpected, as cattle rabies is related to that of D. rotundus and not to that of frugivorous bats.34,35 The most plausible explanation is that these two strains have been transmitted from an Artibeus spp. to a D. rotundus, and then from this bat to the cattle in a rare class of spill-over transmission.
Kobayashi et al.,16 analyzing the N gene of the RABV isolated from frugivorous bats, insectivorous bats, and D. rotundus, reported lineages associated with Artibeus spp. bats (frugivorous), D. rotundus, and insectivorous bats, suggesting that there are species-specific lineages.26 However, they have not provided significant results to distinguish between RABV isolated from D. rotundus and Artibeus spp., possibly due to a restricted number of samples and sampling area. In the present study, this problem was compensated by the inclusion of 35 RABV isolates of bats (20 from Artibeus spp. and 15 from D. rotundus) from a broad geographical area of São Paulo. In addition, this study also analyzed the complete G gene, which provides a more specific distinction between the genetic sequences.
Different hosts pose different challenges for rabies control. This is more complex in bats due to the large number of species, the different ecologic niches that they occupy, and the impossibility to vaccinate these animals. For instance, the population of D. rotundus in Latin America can be legally controlled with vampiricide anticoagulants applied on cattle or on the bats themselves.34 Conversely, non-hematophagous bats are under legal protection, and only now the knowledge of rabies epidemiology in these bats species is increasing.35,36
In this context, the results obtained in this study are valuable, because based on the partial amino acid sequences for the N gene it is possible to differentiate RABV strains from Artibeus spp. and D. rotundus for the purpose of defining the infection sources in molecular epidemiology. These results show the close host relationship of RABV transitions, and have an invaluable application for determining the sources of rabies infections transmitted mainly to dogs and cats in urban centers.
In conclusion, for rabies virus isolates related to frugivorous bats of the Artibeus spp. and to the vampire bat D. rotundus, the phylogeny based on sequences of the N and G genes shows segregation patterns in genus-specific agreement in each of these bats. Data from this study suggest that a lineage of RABV is possibly being established in the Artibeus spp. genus.
Conflict of interestAll authors declare to have no conflict of interest.
The authors are grateful to CAPES (W.O. Fahl's PhD fellowship) and CNPq (P.E. Brandão's PQ-2 fellowship).