To study the potential factors include gene mutation, efflux pump and alteration of permeability associated with quinolone-resistance of Salmonella enterica strains isolated from patients with acute gastroenteritis and to evaluate the degree of synergistic activity of efflux pump inhibitors when combined with ciprofloxacin against resistant isolates.
MethodsAntimicrobial resistance patterns of fifty-eight Salmonella isolates were tested. Five isolates were selected to study the mechanism of resistance associated with quinolone group, including mutation in topoisomerase-encoding gene, altered cell permeability, and expression of an active efflux system. In addition, the combination between antibiotics and efflux pump inhibitors to overcome the microbial resistance was evaluated.
ResultsFive Salmonella isolates totally resistant to all quinolones were studied. All isolates showed alterations in outer membrane proteins including disappearance of some or all of these proteins (Omp-A, Omp-C, Omp-D and Omp-F). Minimum inhibitory concentration values of ciprofloxacin were determined in the presence/absence of the efflux pump inhibitors: carbonyl cyanide m-chlorophenylhydrazone, norepinephrin and trimethoprim. Minimum inhibitory concentration values for two of the isolates were 2–4 fold lower with the addition of efflux pump inhibitors. All five Salmonella isolates were amplified for gyrA and parC genes and only two isolates were sequenced. S. Enteritidis 22 had double mutations at codon 83 and 87 in addition to three mutations at parC at codons 67, 76 and 80 whereas S. Typhimurium 57 had three mutations at codons 83, 87 and 119, but no mutations at parC.
ConclusionsEfflux pump inhibitors may inhibit the major AcrAB-TolC in Salmonella efflux systems which are the major efflux pumps responsible for multidrug resistance in Gram-negative clinical isolates.
Salmonella enterica is a bacterial pathogen that causes a variety of diseases in humans, including gastroenteritis, bacteremia, and typhoid fever.1 Antimicrobial agents are not usually used for the treatment of salmonellosis but can be lifesaving in case of severe or systemic infection.2 Quinolones and fluoroquinolones are broad spectrum antibacterial agents commonly used in both clinical and veterinary medicine. They are a group of synthetic antimicrobial agents with excellent activity against Enterobacteriaceae. However, the number of quinolone-resistant isolates has increased in recent years and such resistance can counteract the efficacy of antimicrobial therapy.3,4 The global increase in the prevalence of Salmonella strains with reduced susceptibility to quinolones constitutes a major concern, since these pathogens have been associated with a significant burden of hospitalization and mortality and with clinical failures of therapy.5–7
Quinolone resistance in Gram-negative pathogens is usually acquired by chromosomal mutations, primarily in the quinolone resistance-determining regions (QRDRs) of the target genes, gyrA and gyrB, which encode DNA gyrase, and parC and parE, which encode topoisomerase IV.8 Quinolones act by inhibiting the action of type II topoisomerases, DNA gyrase and topoisomerase IV.9 Resistance can also be acquired by minimizing the accumulation of the antimicrobial agent in the cell by either one or both of the following mechanisms: altered expression of porins leading to decreased penetration of fluoroquinolones within bacteria or increased efflux of quinolones from the bacterial cell due to overexpression of the AcrAB-TolC efflux pump that acts synergistically with the outer membrane mutation.3,10
Efflux pumps that contribute to resistance to many classes of antimicrobial agents, including fluoroquinolones, have been described in a number of clinically important bacteria such as Salmonella, Escherichia coli, Pseudomonas aeruginosa and others.2 Pumps may be specific for one substrate or may transport a range of structurally dissimilar compounds (including antibiotics of multiple classes); such pumps can be associated with multiple drug resistance (MDR). In the prokaryotic kingdom there are five major families of efflux transporter.2,11 Most of the multidrug transporters belonging to the RND family interact with a membrane fusion protein (MFP) and an outer membrane protein (OMP) to allow drug transport across both the inner and the outer membranes of Gram-negative bacteria.12 The RND efflux pump AcrAB-TolC in Enterobacteriaceae and homologues thereof in other Gram-negative bacteria is the system most commonly associated with innate and acquired chromosomally mediated MDR. Clinical isolates that overexpress efflux pumps usually overproduce this pump and there is an association of MDR mediated by efflux with prior use of fluoroquinolones.4 Due to the lack of new antibacterial agents, there is considerable interest in restoring the activity of older antimicrobial compounds. One way to do this is to inhibit the action of MDR efflux pumps that confer innate resistance to these older agents; this is an area of active drug development by pharmaceutical companies.13 In order to overcome resistance due to overexpressions of efflux pump, efflux pump inhibitors (EPIs) were used by different functions,4 some of these EPIs affect antibiotic efflux not by inhibiting the pump but rather by competing favorably with the antibiotic for extrusion.14
The goal of this study was to determine the resistance mechanism to fluoroquinolones in Salmonella isolates and investigate the association of quinolone resistance with mutations in the genes encoding for DNA gyrase and topoisomerase IV of S. enterica, with the study of quinolones efflux from the bacterial cell and permeability alteration. In addition, to study the use of efflux pump inhibitors to increase the accumulation of fluoroquinolones in Salmonella.
Materials and methodsBacterial isolatesThe bacterial strains used in this study were collected from Clinical Microbiological Laboratory of three hospitals in Cairo, Egypt along a period of eighteen months (from July 2008 through December 2009). One hundred isolates were obtained from a culture of either blood or stool from individual patients with acute gastroenteritis. Salmonella isolates were selected using standard selective and differentiated media, biochemical tests, and serological test based on standard criteria. S. enterica serovar Typhimurium ATCC 14028 was used as a reference strain.
Antibacterial susceptibility testingAntimicrobial resistance patterns of fifty-eight (58) Salmonella isolates selected out of 100 samples were tested, using different antimicrobial agents, by the agar disk diffusion method. Bacteria were grown until they reached a final inoculum of 5×105CFU/mL as compared to 0.5 McFarland standard then streaked on Mueller-Hinton agar (Difco). Used antimicrobial disks (μg/disc) (Oxoid, UK) included ampicillin (AMP, 10μg), oxacillin (OX, 1μg), cefotaxime (CTX, 30μg), ceftazidine (CAZ, 30μg), cefipime (FEP, 30μg), amoxicillin/clavulinic acid (AMC, 30μg), as β-lactam group; levofloxacin (LEV, 5μg), ofloxacin (OFX, 5μg), norfloxacin (NOR, 30μg), ciprofloxacin (CIP, 5μg) and nalidixic acid (NA, 30μg) as fluoroquinolones group and streptomycin (S, 10μg), gentamycin (GN, 10μg) and amikacin (AK, 30μg) as aminoglycosides group. Plates were incubated at 37°C for 24h. Salmonella isolates were defined as sensitive, intermediate or resistant based on the size of the inhibition zone according to breakpoints established by CLSI documents.15
Preparation and analysis of outer membrane proteins (OMPs)Five multidrug resistant, especially to fluoroquinolones group, clinical isolates of S. enterica were selected for genetic study. Outer membrane protein was obtained from an exponential phase culture as described by Miró et al.16 Bacteria were separated by centrifugation (5000×g, 4°C, 15min) and were subsequently sonicated. Cell envelopes were recovered by centrifugation (5000×g, 4°C, 15min) and treated with laurylsarcosynate (Sigma). OMPs were obtained by means of an additional centrifugation step (13,000×g, 4°C, 60min). OMPs extracts were solubilized in sample buffer (final concentration: 10mg/mL) submitted to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and stained using Comassie blue, then the proteins band analyzed using Gel Pro Analyzer V. 3.0 to determine the molecular weights and amount of protein bands.
Detection of active efflux pump systemsA variety of methods has been used to identify active efflux systems in bacteria, such as fluorometric assays or the determination of the minimum inhibitory concentration (MIC) for fluoroquinolones group in the presence of compounds known to modulate the activity of efflux pumps (as efflux pump inhibitors, EPIs)
The Ethidium Bromide-agar Cartwheel (EtBrCW) method proved to be a valuable tool for the rapid and simple screening of efflux pump activity,17 for five clinical isolates of selected MDR S. enterica. Briefly, each culture was swabbed onto TSA plates containing EtBr concentrations ranging from 0.1 to 0.5mg/L (0.1mg/L increments). The plates were incubated at 37°C for 16h, after which the minimum concentration of EtBr associated with the bacterial mass that produced fluorescence under UV light using transilluminator of 312nm (Model LMS-20E, USA) was determined.
To assess the activity of AcrAB efflux pumps, minimal inhibitory concentrations (MICs) of ciprofloxacin were performed by microtiter plate method alone and in the presence of different putative efflux pump inhibitors (EPIs): carbonyl cyanide m-chlorophenylhydrazone (CCCP), norepinephrine (NOR) and trimethoprim (TMP) (Sigma, St. Louis, USA). MICs at concentrations ranging from 0.125 to 512μg/mL were determined for five selected Salmonella isolates. All antibiotics and efflux pump inhibitors were used according to the manufacturer's instructions.
Detection of target gene mutationsFive clinical isolates of S. enterica were used to obtain in vitro FQR and two of them were selected for detection of target gene mutation. Polymerase chain reaction (PCR) amplification was carried out to detect the presence of gyrase and Topo IV enzymes in Salmonella. DNA sequencing detects mutations in the quinolone resistance determining regions (QRDRs) of the gyrA and parC genes. Amplification was carried out using PAPD-PCR cycling program in thermal cyclers (Techne-TC-512 PTC-100:M). The primers (Sigma–Aldrich, USA) were designed according to Eaves et al.,18,19 and were used for PCR amplification and sequencing as listed in Table 1 which includes all bases previously detected as mutation hot spots.
Oligonucleotide primers used for amplification of QRDR for the RAPD PCR assay and sequencing of gyrA and parC genes.
| Primer | Primer (5′–3′) | Amplicon size | Length |
|---|---|---|---|
| gyrA | |||
| Fw | CGTTGGTGACGTAATCGGT | 251bp | 70–152 |
| Rv | CCGTACCGTCATAGTTATC | ||
| parC | |||
| Fw | CTATGCGATG TCAGAGCTGG | 270bp | 47–133 |
| Rv | TAACAGCAGCTCGGCGTATT | ||
Fw, forward; Rv, reversible; bp, base pair.
PCR products were extracted and purified using DNA gel extraction kit (Montage, Millipore) and sequencing was performed by Jena Gen GmbH Biotechnologie-Gentechnik-Diagnostik (Jena, Germany) using the BigDye™ Cycle Sequencing Kit (Applied Biosystems, Weiterstadt, Germany) on a 3130 sequencer (Applied Biosystems).
The results were analyzed using Bioedit software version 7.0 where the nucleotides of gyrA and parC from QRDR were compared to nucleotides of Salmonella isolates in GenBank accession nos. X78977 and AE008878, respectively.
ResultsFifty-eight (58%) out of the 100 isolates collected from the three hospitals were identified by microscopic examination, selective and differential media, biochemical tests and serological test as S. enterica. Out of the 58% Salmonella isolates, 41.3% represented S. Typhimurium, 27.58% S. Enteritidis, 5.17% S. Typhi, 3.44% both S. Paratyphi A and B, and 18.96% other Salmonella. All Salmonella isolates were assayed for resistance to antibacterial agents of three groups. Only five (8.62%) among the tested strains appeared to be totally resistant to all antibacterial agents of the quinolone and fluoroquinolone groups and to most of the agents of the two other groups (Table 2). The five resistant isolates were S. enterica serovar Typhimurium number 7, 54 and 57 (S-7, S-54 and S-57) and S. enterica serovar Enteritidis number 22 (S-22) and S. Typhi number 49 (S-49).
Antimicrobial susceptibility of selected five multidrug resistant Salmonella isolates according to CLSI (2012).
| Antibiotics | Salmonella isolates | ||||
|---|---|---|---|---|---|
| S. Tm 7 | S. En 22 | S. Ty 49 | S. Tm 54 | S .Tm 57 | |
| Aminoglycosides | |||||
| Streptomycin (S) | R | S | R | R | R |
| Gentamycin (GN) | R | R | R | R | R |
| Amikacin (AK) | R | R | R | R | I |
| β-Lactams | |||||
| Ampicillin (AMP) | R | R | R | R | R |
| Amoxicillin/clavulanic acid (AMC) | R | R | R | R | R |
| Oxacillin (OX) | R | R | R | R | R |
| Cefotaxime (CTX) | R | R | I | R | R |
| Ceftazidine (CAZ) | R | R | S | R | S |
| Cefipime (FEP) | R | R | R | R | R |
| Quinolones and fluoroquinolones | |||||
| Ciprofloxacin (CIP) | R | R | R | R | R |
| Levofloxacin (LEV) | R | R | R | R | R |
| Ofloxacin (OFX) | R | R | R | R | R |
| Norfloxacin (NOR) | R | R | R | R | I |
| Nalidixic acid (NA) | R | R | R | R | R |
Susceptible, intermediate and resistant phenotypes, from CLSI breakpoints.
In the present study OMP expression was altered in the five multidrug resistant Salmonella isolates, based on electrophoretic mobility (Table 3), and the reference strain S. Typhimurium ATCC 14028 had OmpA, OmpC, OmpD and OmpF. Expression of Omp-F was lost in all isolates except in S. Typhimurium (S-7), while OmpD was expressed in S. Typhimurium (S-57) only. The isolates S. Typhimurium (S-54) and S. Entreritidis (S-22) did not show the major non-specific outer membrane proteins. Quinolone MIC for the former isolate was higher than for the other four isolates. Salmonella Typhi (S-49) had lower quinolone MIC than the four other isolates (Table 3); it had two bands corresponding to OmpA and OmpC. Also, the major non-specific outer membrane protein with the molecular weight of 49kDa was detected in all multidrug resistant Salmonella isolates.
Major OMP of five multidrug resistant Salmonella isolates.
| Isolate no. | Serovar | MICs (μg/mL) | Protein content (μg/mL) | |||
|---|---|---|---|---|---|---|
| OmpA | OmpC | OmpD | OmpF | |||
| Sensitive strain | S. Typhimurium ATCC 14028 | – | 6.52 | 5.35 | 5.22 | 6.64 |
| S-7 | S. Typhimurium | 64 | – | – | – | 3.16 |
| S-22 | S. Enteritidis | 256 | – | – | – | – |
| S-49 | S. Typhi | 32 | 2.23 | 2.89 | – | – |
| S-54 | S. Typhimurium | >512 | – | – | – | – |
| S-57 | S. Typhimurium | 512 | – | – | 1.51 | – |
MIC, minimum inhibitory concentration; Omp, outer membrane protein.
The application of EtBrCW method allowed for the selection of two EtBrCW-positive (isolates S-22 and S-57) isolates, whereas the remaining three isolates were considered to have no or intermediate efflux activity. Based upon these results, we continued the study by further analyzing the two EtBrCW-positive isolates.
Three compounds of efflux pump inhibitors (EPI) were used to decrease the MICCIP by two different mechanisms: carbonyl cyanide m-chlorophenylhydrazone (CCCP) (inhibition of proton-motive force driven efflux pumps),21,22 norepinephrine (NOR) and trimethoprim(TMP) (inhibits specific RND efflux pumps) and is known to be active against the AcrAB-TolC transporters.4 MICCIP determined in absence of EPIs were compared with those determined in presence of efflux inhibitors. The results showed twofold or more reduction in MIC levels (Table 4) for the two multidrug resistant Salmonella isolates (EtBrCW-positive isolates) tested.
MICs values of different EPIs used against quinolone resistant Salmonella isolates and molecular characterization of QRDR region of gyrA and parC gene.
| Isolate no. | Serovar | QRDR mutation | MICs (μg/mL) | ||||
|---|---|---|---|---|---|---|---|
| CIP | |||||||
| gyrA | parC | NOEPI | +CCCP | +NOR | +TMP | ||
| S22 | S. Enteritidis | Ser83PheAsp87Ser | Ser67CysArg76CysCys80Arg | 256 | 32 (−3) | 64 (−2) | 16 (−4) |
| S57 | S. Typhimurium | Ser83PheAsp87GlyAla119Ser | No mutation | 512 | 128 (−2) | 32 (−4) | 64 (−3) |
Amino acids: serine (Ser), phenylalanine (Phe), aspartic acid (Asp), glycine (Gly), arginine (Arg), alanine (Ala), cysteine (Cys), ciprofloxacin (CIP), carbonyl cyanide m-chlorophenylhydrazone (CCCP), norepinephrine (NOR), trimethoprim (TMP), efflux pump inhibitor (EPI), quinolone resistance determining region (QRDR).
Numbers in parenthesis represent the numbers of fold decrease.
As ciprofloxacin is a substrate of many efflux pumps in different bacterial species,23 the study focused upon usage of compounds that displayed synergistic activity with ciprofloxacin. So the use of the EPIs like CCCP, trimethoprim and norepinephrine in combination with antibiotic revealed synergistic antibacterial activity for improving and potentiating the activity of antibiotics.
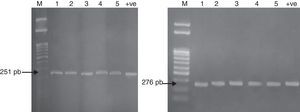
The mechanism of resistance to fluoroquinolones is caused by mutations in a region of the gyrA gene resulting in amino acid changes between amino acid positions 67 and 106. This region has been termed the quinolone resistance-determining region (QRDR). The QRDR of gyrA (251bp) and parC (276bp) was amplified for the five multidrug resistant Salmonella isolates as represented in Fig. 1 to detect the presence of these genes and confirm the identification of the five Salmonella isolates.
The QRDR of the genes gyrA and parC of two selected QR Salmonella isolates (S22 and S57) were tested for mutations by DNA amplification and sequencing. Results revealed that S. Enteritidis (S-22) and S. Typhimurium (S-57) contained double mutation at codon 83 (Ser83-Phe) and codon 87 (Asp87-Ser in S22 or Gly in S57). One novel mutation was detected in S. Typhimurium (S-57) inside QRDR, Ala-119, in which G-T led to the substitution of alanine to serine (Ala119-Ser); while in S. Enteritidis (22) there is seven mutations within QRDR from codon (115–122) and other mutations outside QRDR. The 251-nucletide fragment of gyrA and 270 nucleotide fragment of parC that were amplified from the quinolone-resistant S. Typhimurium (S-57) and S. Enteritidis (S-22) had over 70% and 50% identity with the same fragment from the reference strain (X78977) and reference strain (AE008878), respectively. Other mutations found in different positions did not seem to have clinical significance.
DiscussionThe occurrence of antibiotic-resistant bacteria of all genera increased recently. The scientific community should, therefore, perform routine surveillance of microbial population to determine the extent of antibiotic resistance in order to provide suitable treatment guidelines.
Davin-Regli et al. and O’Regan et al. found that the loss of one OMPs or the decrease in its amount compared with the parent strain cause a high resistance rate in Salmonella isolates.25,26 This was also seen here in the five tested Salmonella isolates. Miró et al. discussed the relevance of protein losses to resistance to β-lactams and quinolones, stating that the expression of the other porin presumably allows for sufficient penetration of antimicrobial agents to inhibit the microorganism as shown in Klebsiella pneumoniae and E. coli.16
Ruiz et al. stated that analysis of OMPs and lipopolysaccharides from quinolone-susceptible and quinolone-resistant clinical isolates showed no porin changes.27 Hamid and Jain reported that isolates which contain the non-specific outer membrane protein with the molecular weight of 49KDa may be used as a vaccine for protection against typhoid.28
Efflux pumps are transport proteins involved in the extrusion of toxic substrates (including virtually all classes of clinically relevant antibiotics) from within cells into the external environment.29 Efflux activity was assessed by means of a fast and practical test, the Ethidium Bromide-agar Cartwheel (EtBrCW), which provides information on the capacity of each isolate to extrude EtBr from the cells by efflux, on the basis of the fluorescence emitted by cultures swabbed in EtBr-containing agar plates.
The outer membrane modification acts synergistically with enhanced active efflux systems to affect the level of the intrinsic and the acquired antibiotic resistance of Gram-negative bacteria as stated by Nikaido.30 The smallest concentration of EtBr that produces fluorescence of the bacterial mass represents the highest concentration of EtBr that the bacteria can extrude, consequently producing fluorescence is significantly greater. The concentration of EtBr needed to produce fluorescence of the bacterial mass is considerably higher than that concentration which produces fluorescence of the wild-type strain.31
Several chemical families of EPIs have now been described and characterized.20 Among them several inhibitor compounds display an efficient activity and inhibit the major AcrAB-TolC in Salmonella efflux systems which are the major efflux pumps responsible for MDR Gram-negative clinical isolates. The NOR and TMP have structure analog as phenyl-arginine-β-naphthylamide (PAβN). PAβN is routinely used in the laboratory as a screen to indicate efflux-mediated antibiotic resistance in Gram-negative bacteria. However, PAβN is not used in the clinical setting due to toxicity and bioavailability issues, but NOR and TMP are already licensed for use in man.4,32
The twofold or greater reduction in MIC levels obtained here for the two multidrug resistant Salmonella isolates confirm the presence of an active efflux component in those isolates.4,33 Piddock et al.4 found the MIC values of ciprofloxacin to be 2–4 fold reduced in the presence of trimethoprim for wild-type strains of Salmonella Typhimurium and NCTC type strains of Enterobacter cloacae, Serratia marcescens, P. aeruginosa, K. pneumoniae and E. coli. However, no synergy was seen between trimethoprim and ciprofloxacin for strains of Salmonella Typhimurium in which acrA, acrB, acrAB or tolC were inactivated. Also Keddy et al. found that in the presence of EPIs, the MIC of ciprofloxacin decreased by twofold, and the MIC of nalidixic acid decreased by fivefold, establishing the involvement of an efflux pump in conferring quinolone resistance.34 TMP and NOR were the most effective inhibitors in reducing MIC values. While CCCP showed less effect on MIC values for the compounds tested, this result provided evidence for an active efflux process that uses the proton motive force as energy source.21,22 However, no full reversion of the ciprofloxacin resistance phenotype was obtained with any of the EPIs tested, suggesting the contribution of other mechanisms to this resistance, namely, mutations in the target genes.
The main mechanism of resistance to fluoroquinolones in Gram-negative bacteria is caused by mutations in genes coding for DNA-gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE).24 Results of the test for mutations, by DNA amplification and sequencing the QRDR of the genes gyrA and parC of two selected QR Salmonella isolates (S22 and S57) agree with those of Guerra et al.,35 Marimón et al.,33 and Chen et al.2 Griggs et al. suggested that other mutations could be present in the gyrA genes of these isolates, outside of the region of the sequenced gene, and that the Ala119 mutation is not involved in quinolone resistance.36 Eaves et al. found novel mutations inside QRDR at codon Asp72, Asp82 and Ala119 and also outside the QRDR, but within the amplified region of DNA.18
Amino acid changes at Ser-83 (to Phe, Tyr or Ala) or at Asp-87 (to Gly, Asn or Tyr) are the most frequently observed in nalidixic acid-resistant strains.37 Double mutations at both residues 83 and 87 have been identified in clinical isolates of S. Typhimurium DT204 showing high-level resistance to fluoroquinolones.35 Nonetheless, O’Regan et al. stated that there is a single gyrA mutation (D87Y) present in all strains showing high-level nalidixic acid resistance.26 Hirose et al.39 and Dimitrove et al.38 demonstrated that single point mutation in QRDR of gyrA led to decreased susceptibility to ciprofloxacin in Salmonella isolates. There is also three amino acid substitution in parC at codon Ser67-Cys, Arg76-Cys and Cys80-Arg in S. Enteritidis (S-22), in agreement with results obtained by Choi et al. and Wu et al.40,41 However, in S. Typhimurium S-57 and the in wild strains no amino acid mutation in parC gene has been reported. It is unlikely that this mutation contributes to quinolone resistance. This indicates that this substitution is likely to be a polymorphism not common in resistant strains of Salmonella.
Obtained data showed that S. Typhimurium (S-57) has high MICCIP with mutations in only gyrA, while S. Entridies (S-22) has lower MICCIP with mutation in gyrA and parC. These results clearly suggest that parC mutations are not necessary to generate high level resistance to ciprofloxacin. In this respect Eaves et al.19 found that isolates with a mutation in both gyrA and parC were more susceptible to ciprofloxacin than were isolates with a mutation in gyrA alone. Previous studies42 have demonstrated that changes in topoisomerases are insufficient to explain the level of resistance to quinolones in S. enterica. In agreement with this observation, Miró et al.16 showed that multiple resistance mechanisms were involved in the strains including changes in topoisomerases, altered permeability, and expression of an active efflux. Our results showed that multiple resistance mechanisms were involved in the strains including changes in topoisomerases, as shown in double mutation at both codons 83 and 87 in gene gyrA and mutation inside QRDR in both isolates and three mutations in parC only in S. Enteritidis, altered permeability, and expression of an active efflux.
ConclusionThe data obtained in this work strongly suggest the importance of mutations in gyrase and topoisomerase in quinolone resistance development in Salmonella, although other factors such as overexpression of efflux pumps can play a complementary role. Additionally, our study underscores the importance of using EPI (NOR and TMP) to inhibit the major AcrAB-TolC in Salmonella efflux systems which are the major efflux pumps responsible for MDR Gram-negative clinical isolates. MDR and fluoroquinolone-resistance in Salmonella recovered from patients are common in Egypt, and monitoring antimicrobial resistance of foodborne pathogens should be an important component of the surveillance system to improve public health.
Conflict of interestThe authors declare no conflict of interest.