This study evaluated the in vitro activity of ceftolozane-tazobactam and comparator agents tested against Latin American isolates of Enterobacteriaceae and Pseudomonas aeruginosa from patients with health care-associated infections. Ceftolozane-tazobactam is an antipseudomonal cephalosporin combined with a well-established β-lactamase inhibitor.
A total of 2415 Gram-negative organisms (537 P. aeruginosa and 1878 Enterobacteriaceae) were consecutively collected in 12 medical centers located in four Latin American countries. The organisms were tested for susceptibility by broth microdilution methods as described by the CLSI M07-A10 document and the results interpreted according to EUCAST and CLSI breakpoint criteria.
ResultsCeftolozane-tazobactam (MIC50/90, 0.25/32μg/mL; 84.2% susceptible) and meropenem (MIC50/90, ≤0.06/0.12μg/mL; 92.6% susceptible) were the most active compounds tested against Enterobacteriaceae. Among the Enterobacteriaceae isolates tested, 6.6% were carbapenem-resistant Enterobacteriaceae and 26.4% exhibited an extended-spectrum β-lactamase non-carbapenem-resistant phenotype. Whereas ceftolozane-tazobactam showed good activity against extended-spectrum beta-lactamase, non-carbapenem-resistant phenotype strains of Enterobacteriaceae (MIC50/90, 0.5/>32μg/mL), it lacked useful activity against strains with a (MIC50/90, >32/>32μg/mL; 1.6% S) carbapenem-resistant phenotype. Ceftolozane-tazobactam was the most potent (MIC50//90, 0.5/16μg/mL) β-lactam agent tested against P. aeruginosa isolates, inhibiting 86.8% at an MIC of ≤4μg/mL. P. aeruginosa exhibited high rates of resistance to cefepime (16.0%), ceftazidime (23.6%), meropenem (28.3%), and piperacillin-tazobactam (16.4%).
ConclusionsCeftolozane-tazobactam was the most active β-lactam agent tested against P. aeruginosa and demonstrated higher in vitro activity than available cephalosporins and piperacillin-tazobactam when tested against Enterobacteriaceae.
The epidemiology of microbial pathogens causing health care-associated infections (HAIs) has changed dramatically over the last decades with a concomitant increase in antibiotic resistance.1–5 Whereas resistant Gram-positive cocci (GPC) were a major concern during the 1990s,1,6 more recently multidrug-resistant (MDR; resistant to ≥3 classes of agents) Gram-negative bacilli (GNB) have become increasingly prevalent in the hospital setting.1–4 This is especially true in Latin American countries where MDR-GNB, such as Pseudomonas aeruginosa, carbapenem-resistant (CRE), and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae are a serious threat.2–4 Due to the relative lack of new agents to treat these infections,7 empirical therapy is often ineffective and requires combinations of antibacterial agents to achieve optimal coverage.2,8
These findings underscore the continued importance of antibiotic resistance surveillance and the need to assess the potential impact of newly introduced and novel antibacterial agents targeting specific resistance phenotypes.9,10 Systematic and comprehensive antibiotic resistance surveillance is essential to document the extent of the resistance problem and to inform local, regional, national, and global efforts to combat the resistance challenge.9 The SENTRY Antimicrobial Surveillance Program has monitored the predominant pathogens and antimicrobial resistance patterns of HAI pathogens via a network of sentinel sites in Latin America since 1997 and has documented the steady emergence of MDR-GNB in those countries.2,3,6,11
Ceftolozane-tazobactam is a novel antibacterial agent with activity against P. aeruginosa, including antibiotic-resistant strains, and other common GNB, including most ESBL-producing Enterobacteriaceae strains.10,12–15 Ceftolozane-tazobactam has limited activity against Acinetobacter spp.; Stenotrophomonas maltophilia; GPC; organisms producing carbapenemases or metallo-β-lactamases; or a minority of AmpC β-lactamases found in Enterobacteriaceae.10,16 Ceftolozane-tazobactam was recently approved to treat complicated intra-abdominal infections (cIAI) and complicated urinary tract infections (cUTI).10 A Phase 3 clinical trial of ceftolozane-tazobactam to treat nosocomial pneumonia is ongoing.
In 2012 North American and European antimicrobial resistance surveys, we described the in vitro activity of ceftolozane-tazobactam tested against isolates of Enterobacteriaceae and P. aeruginosa from different infection sites.13,14 In this study, we extended those observations and focused on the activity of ceftolozane-tazobactam and comparators against 2415 isolates collected from 2013 through 2015 comprising P. aeruginosa (537 isolates) and Enterobacteriaceae (1878 isolates) from patients with HAIs hospitalized at 12 Latin American medical centers (four countries). The analysis includes the activity of ceftolozane-tazobactam against specific resistant phenotypes (e.g., ESBL non-CRE phenotype and MDR strains of Enterobacteriaceae and P. aeruginosa) as well as the frequencies of resistance phenotypes among the Latin American countries.
Materials and methodsSampling sites and organismsA total of 2415 non-duplicate isolates of GNB, including 1878 Enterobacteriaceae and 537 P. aeruginosa, were consecutively collected in 12 medical centers in four Latin American countries between January 1, 2013, and December 31, 2015. Each participating medical center identified species that were confirmed by the monitoring laboratory (JMI Laboratories, North Liberty, Iowa, USA) using the VITEK 2 System (bioMérieux, Hazelwood, Missouri, USA) or matrix-assisted laser desorption ionization-time of flight mass spectrometry (Bruker, Billerica, Massachusetts, USA), when necessary.
Antimicrobial susceptibility testingMinimal inhibitory concentrations (MICs) were determined using the reference Clinical and Laboratory Standards Institute (CLSI) broth microdilution method.17 Quality control (QC) and interpretation of results were performed in accordance with CLSI M100-S26 and European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2016 guidelines.18,19Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Proteus mirabilis were grouped as “ESBL phenotype” based on the CLSI screening criteria for potential ESBL production, i.e., MIC of ≥2μg/mL of ceftazidime, ceftriaxone, or aztreonam.18 CRE were defined as isolates displaying MIC values of ≥4μg/mL18 for imipenem (P. mirabilis and indole-positive Proteeae were not included due to the intrinsically elevated MIC values), meropenem, and/or doripenem. Since carbapenemase-producing isolates may also appear to have an ESBL phenotype, non-carbapenem-resistant ESBL-phenotype isolates were analyzed (ESBL non-CRE). P. aeruginosa isolates were classified as ceftazidime nonsusceptible (NS; MIC, >8μg/mL) and meropenem-NS (MIC, >2μg/mL).
TheoryWe performed surveillance of antimicrobial resistance among isolates of Enterobacteriaceae and P. aeruginosa from Latin America. The analysis includes the activity of ceftolozane-tazobactam against specific resistant phenotypes as well as the frequencies of occurrence of the resistance phenotypes among the Latin American countries. The percent susceptible to ceftolozane-tazobactam as well as the MICs encompassing 50% and 90% of the tested populations are presented.
ResultsThe countries contributing isolates (sites; sample size) were: Argentina (2; 486), Brazil (5; 869), Chile (2; 471), and Mexico (3; 709). All organisms were isolated from hospitalized patients with bloodstream infections (BSI; 840 isolates), hospital-associated pneumonia (639 isolates), skin and skin structure infections (799 isolates), and other infections (137 isolates).
Among the 2415 isolates tested were 1878 Enterobacteriaceae isolates (including 661 E. coli isolates, 653 Klebsiella spp. isolates, 250 Enterobacter spp. isolates, 33 Citrobacter spp. isolates, 100 P. mirabilis isolates, 60 indole-positive Proteeae isolates, and 107 Serratia marcescens isolates) and 537 P. aeruginosa isolates (Table 1).
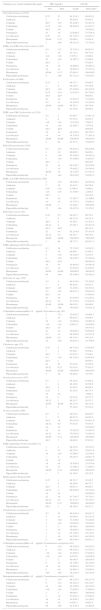
Antimicrobial activity of ceftolozane-tazobactam tested against the main organisms and organism groups of isolates (μg/mL).
| Organism/organism group | No. of isolates at MIC (μg/mL; cumulative %) | MIC50 | MIC90 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | > | |||
| Enterobacteriaceae(1878) | 1 (0.1%) | 1 (0.1%) | 20 (1.2%) | 343 (19.4%) | 589 (50.8%) | 421 (73.2%) | 145 (80.9%) | 61 (84.2%) | 33 (85.9%) | 40 (88.1%) | 30 (89.7%) | 38 (91.7%) | 156 (100.0%) | 0.25 | 32 |
| CRE (124) | 0 (0.0%) | 1 (0.8%) | 0 (0.8%) | 0 (0.8%) | 0 (0.8%) | 0 (0.8%) | 1 (1.6%) | 0 (1.6%) | 2 (3.2%) | 8 (9.7%) | 12 (19.4%) | 21 (36.3%) | 79 (100.0%) | >32 | >32 |
| ESBL non-CRE (495) | 0 (0.0%) | 1 (0.2%) | 12 (2.6%) | 83 (19.4%) | 152 (50.1%) | 83 (66.9%) | 39 (74.7%) | 19 (78.6%) | 18 (82.2%) | 15 (85.3%) | 14 (88.1%) | 59 (100.0%) | 0.5 | >32 | |
| Escherichia coli(661) | 0 (0.0%) | 14 (2.1%) | 195 (31.6%) | 265 (71.7%) | 113 (88.8%) | 41 (95.0%) | 11 (96.7%) | 4 (97.3%) | 3 (97.7%) | 7 (98.8%) | 2 (99.1%) | 6 (100.0%) | 0.25 | 1 | |
| Non-ESBL phenotype (421) | 0 (0.0%) | 14 (3.3%) | 187 (47.7%) | 208 (97.1%) | 12 (100.0%) | 0.25 | 0.25 | ||||||||
| ESBL phenotype (240) | 0 (0.0%) | 8 (3.3%) | 57 (27.1%) | 101 (69.2%) | 41 (86.2%) | 11 (90.8%) | 4 (92.5%) | 3 (93.8%) | 7 (96.7%) | 2 (97.5%) | 6 (100.0%) | 0.5 | 2 | ||
| ESBL non-CRE (238) | 0 (0.0%) | 8 (3.4%) | 57 (27.3%) | 101 (69.7%) | 41 (87.0%) | 11 (91.6%) | 4 (93.3%) | 3 (94.5%) | 6 (97.1%) | 2 (97.9%) | 5 (100.0%) | 0.5 | 2 | ||
| Meropenem-susceptible (≤1μg/mL) (659) | 0 (0.0%) | 14 (2.1%) | 195 (31.7%) | 265 (71.9%) | 113 (89.1%) | 41 (95.3%) | 11 (97.0%) | 4 (97.6%) | 3 (98.0%) | 6 (98.9%) | 2 (99.2%) | 5 (100.0%) | 0.25 | 1 | |
| Klebsiellaspp. (653) | 0 (0.0%) | 1 (0.2%) | 6 (1.1%) | 99 (16.2%) | 172 (42.6%) | 85 (55.6%) | 48 (62.9%) | 27 (67.1%) | 15 (69.4%) | 22 (72.7%) | 20 (75.8%) | 30 (80.4%) | 128 (100.0%) | 0.5 | >32 |
| Klebsiella pneumoniae (594) | 0 (0.0%) | 1 (0.2%) | 6 (1.2%) | 79 (14.5%) | 153 (40.2%) | 72 (52.4%) | 48 (60.4%) | 25 (64.6%) | 12 (66.7%) | 22 (70.4%) | 18 (73.4%) | 30 (78.5%) | 128 (100.0%) | 0.5 | >32 |
| Non-ESBL phenotype (254) | 0 (0.0%) | 5 (2.0%) | 75 (31.5%) | 131 (83.1%) | 31 (95.3%) | 11 (99.6%) | 1 (100.0%) | 0.25 | 0.5 | ||||||
| ESBL phenotype (340) | 0 (0.0%) | 1 (0.3%) | 1 (0.6%) | 4 (1.8%) | 22 (8.2%) | 41 (20.3%) | 37 (31.2%) | 24 (38.2%) | 12 (41.8%) | 22 (48.2%) | 18 (53.5%) | 30 (62.4%) | 128 (100.0%) | 16 | >32 |
| CRE (114) | 0 (0.0%) | 1 (0.9%) | 0 (0.9%) | 0 (0.9%) | 0 (0.9%) | 0 (0.9%) | 1 (1.8%) | 0 (1.8%) | 1 (2.6%) | 8 (9.6%) | 11 (19.3%) | 18 (35.1%) | 74 (100.0%) | >32 | >32 |
| ESBL non-CRE (226) | 0 (0.0%) | 1 (0.4%) | 4 (2.2%) | 22 (11.9%) | 41 (30.1%) | 36 (46.0%) | 24 (56.6%) | 11 (61.5%) | 14 (67.7%) | 7 (70.8%) | 12 (76.1%) | 54 (100.0%) | 2 | >32 | |
| Meropenem-susceptible (≤1μg/mL) (465) | 0 (0.0%) | 6 (1.3%) | 79 (18.3%) | 153 (51.2%) | 72 (66.7%) | 47 (76.8%) | 25 (82.2%) | 11 (84.5%) | 13 (87.3%) | 7 (88.8%) | 12 (91.4%) | 40 (100.0%) | 0.25 | 32 | |
| Meropenem-nonsusceptible (>1μg/mL) (129) | 0 (0.0%) | 1 (0.8%) | 0 (0.8%) | 0 (0.8%) | 0 (0.8%) | 0 (0.8%) | 1 (1.6%) | 0 (1.6%) | 1 (2.3%) | 9 (9.3%) | 11 (17.8%) | 18 (31.8%) | 88 (100.0%) | >32 | >32 |
| Klebsiella oxytoca (54) | 0 (0.0%) | 19 (35.2%) | 18 (68.5%) | 12 (90.7%) | 0 (90.7%) | 2 (94.4%) | 1 (96.3%) | 0 (96.3%) | 2 (100.0%) | 0.25 | 0.5 | ||||
| Non-ESBL phenotype (43) | 0 (0.0%) | 19 (44.2%) | 16 (81.4%) | 8 (100.0%) | 0.25 | 0.5 | |||||||||
| ESBL phenotype (11) | 0 (0.0%) | 2 (18.2%) | 4 (54.5%) | 0 (54.5%) | 2 (72.7%) | 1 (81.8%) | 0 (81.8%) | 2 (100.0%) | 0.5 | 16 | |||||
| Enterobacterspp. (250) | 0 (0.0%) | 25 (10.0%) | 85 (44.0%) | 72 (72.8%) | 16 (79.2%) | 12 (84.0%) | 5 (86.0%) | 10 (90.0%) | 3 (91.2%) | 6 (93.6%) | 16 (100.0%) | 0.5 | 8 | ||
| Ceftazidime-susceptible (≤4μg/mL) (165) | 0 (0.0%) | 25 (15.2%) | 83 (65.5%) | 52 (97.0%) | 5 (100.0%) | 0.25 | 0.5 | ||||||||
| Ceftazidime-nonsusceptible (>4μg/mL) (85) | 0 (0.0%) | 2 (2.4%) | 20 (25.9%) | 11 (38.8%) | 12 (52.9%) | 5 (58.8%) | 10 (70.6%) | 3 (74.1%) | 6 (81.2%) | 16 (100.0%) | 2 | >32 | |||
| Enterobacter cloacae species complex (216) | 0 (0.0%) | 22 (10.2%) | 68 (41.7%) | 66 (72.2%) | 14 (78.7%) | 7 (81.9%) | 5 (84.3%) | 9 (88.4%) | 3 (89.8%) | 6 (92.6%) | 16 (100.0%) | 0.5 | 32 | ||
| Enterobacter aerogenes (34) | 0 (0.0%) | 3 (8.8%) | 17 (58.8%) | 6 (76.5%) | 2 (82.4%) | 5 (97.1%) | 0 (97.1%) | 1 (100.0%) | 0.25 | 2 | |||||
| Citrobacter spp. (33) | 0 (0.0%) | 12 (36.4%) | 7 (57.6%) | 2 (63.6%) | 0 (63.6%) | 2 (69.7%) | 4 (81.8%) | 3 (90.9%) | 0 (90.9%) | 0 (90.9%) | 3 (100.0%) | 0.25 | 8 | ||
| Citrobacter koseri (9) | 0 (0.0%) | 7 (77.8%) | 2 (100.0%) | 0.12 | – | ||||||||||
| Citrobacter freundii species complex (24) | 0 (0.0%) | 5 (20.8%) | 5 (41.7%) | 2 (50.0%) | 0 (50.0%) | 2 (58.3%) | 4 (75.0%) | 3 (87.5%) | 0 (87.5%) | 0 (87.5%) | 3 (100.0%) | 0.5 | >32 | ||
| Serratia marcescens (107) | 1 (0.9%) | 0 (0.9%) | 0 (0.9%) | 0 (0.9%) | 13 (13.1%) | 62 (71.0%) | 22 (91.6%) | 4 (95.3%) | 2 (97.2%) | 0 (97.2%) | 0 (97.2%) | 0 (97.2%) | 3 (100.0%) | 0.5 | 1 |
| Proteus mirabilis (100) | 0 (0.0%) | 2 (2.0%) | 23 (25.0%) | 63 (88.0%) | 8 (96.0%) | 2 (98.0%) | 1 (99.0%) | 1 (100.0%) | 0.5 | 1 | |||||
| Non-ESBL phenotype (83) | 0 (0.0%) | 2 (2.4%) | 21 (27.7%) | 58 (97.6%) | 2 (100.0%) | 0.5 | 0.5 | ||||||||
| ESBL phenotype (17) | 0 (0.0%) | 2 (11.8%) | 5 (41.2%) | 6 (76.5%) | 2 (88.2%) | 1 (94.1%) | 1 (100.0%) | 1 | 4 | ||||||
| Indole-positive Proteeae (60) | 0 (0.0%) | 8 (13.3%) | 23 (51.7%) | 16 (78.3%) | 9 (93.3%) | 2 (96.7%) | 1 (98.3%) | 1 (100.0%) | 0.25 | 1 | |||||
| Pseudomonas aeruginosa(537) | 0 (0.0%) | 1 (0.2%) | 4 (0.9%) | 53 (10.8%) | 237 (54.9%) | 93 (72.3%) | 48 (81.2%) | 30 (86.8%) | 15 (89.6%) | 13 (92.0%) | 8 (93.5%) | 35 (100.0%) | 0.5 | 16 | |
| Ceftazidime-susceptible (≤8μg/mL) (376) | 0 (0.0%) | 1 (0.3%) | 4 (1.3%) | 53 (15.4%) | 232 (77.1%) | 69 (95.5%) | 11 (98.4%) | 5 (99.7%) | 1 (100.0%) | 0.5 | 1 | ||||
| Ceftazidime-nonsusceptible (>8μg/mL) (161) | 0 (0.0%) | 5 (3.1%) | 24 (18.0%) | 37 (41.0%) | 25 (56.5%) | 14 (65.2%) | 13 (73.3%) | 8 (78.3%) | 35 (100.0%) | 4 | >32 | ||||
| Meropenem-susceptible (≤2μg/mL) (345) | 0 (0.0%) | 1 (0.3%) | 4 (1.4%) | 51 (16.2%) | 199 (73.9%) | 51 (88.7%) | 24 (95.7%) | 9 (98.3%) | 3 (99.1%) | 0 (99.1%) | 1 (99.4%) | 2 (100.0%) | 0.5 | 2 | |
| Meropenem-nonsusceptible (>2μg/mL) (192) | 0 (0.0%) | 2 (1.0%) | 38 (20.8%) | 42 (42.7%) | 24 (55.2%) | 21 (66.1%) | 12 (72.4%) | 13 (79.2%) | 7 (82.8%) | 33 (100.0%) | 2 | >32 | |||
During 2013–2015, ceftolozane-tazobactam maintained a consistent and potent level of activity against the target pathogens from the Latin American study sites (Table 1). Among the Enterobacteriaceae isolates tested, 6.6% were CRE and 26.4% exhibited an ESBL non-CRE phenotype (Table 1). An ESBL non-CRE phenotype was observed in 36.0% of E. coli and 38.0% of K. pneumoniae isolates. An ESBL phenotype was detected in 20.4% of K. oxytoca and 17.0% of P. mirabilis isolates. Important resistant phenotypes among the P. aeruginosa isolates included ceftazidime-NS (30.0%) and meropenem-NS (35.8%) (Table 1).
Ceftolozane-tazobactam MIC values ranged from ≤0.015 to >32μg/mL, and 84.2% of the tested Enterobacteriaceae isolates were inhibited at an MIC value of ≤2μg/mL (85.9% at ≤4μg/mL) (Table 1). Whereas ceftolozane-tazobactam showed good activity against ESBL non-CRE phenotype Enterobacteriaceae strains (MIC50/90, 0.5/>32μg/mL; 74.7% S), it lacked useful activity against strains with a CRE (MIC50/90, >32/>32μg/mL; 1.6% S) phenotype.
Ceftolozane-tazobactam MIC values ranged from 0.06μg/mL to >32μg/mL against P. aeruginosa isolates, and 89.6% of the tested isolates were inhibited at an MIC value of ≤8μg/mL (86.8% susceptible at the ≤4μg/mL CLSI/EUCAST breakpoint). Among the two resistant phenotypes, 56.5% (ceftazidime-NS) and 66.1% (meropenem-NS) were susceptible to ceftolozane-tazobactam (Table 1).
Activities of ceftolozane-tazobactam and comparators against EnterobacteriaceaeCeftolozane-tazobactam (MIC50/90, 0.25/32μg/mL) inhibited 84.2% of the 1878 Enterobacteriaceae isolates tested at ≤2μg/mL and 85.9% of the isolates at ≤4μg/mL (CLSI susceptible and intermediate breakpoint criteria, respectively) (Tables 1 and 2). Enterobacteriaceae isolates displayed susceptibility rates to other β-lactam agents ranging from 65.2/63.4% for cefepime, 66.8/61.8% for ceftazidime, 80.2/75.7% for piperacillin-tazobactam, and 92.6/93.6% for meropenem using CLSI/EUCAST breakpoints. Using MIC50 values, ceftolozane-tazobactam was equal in potency to that of ceftazidime (MIC50, 0.25μg/mL), was up to 2-fold more active than cefepime (MIC50, ≤0.5μg/mL), was 8-fold more potent than piperacillin-tazobactam (MIC50, 2μg/mL), and was at least 4-fold less potent than meropenem (MIC50, ≤0.06μg/mL) (Table 2). Among the non-β-lactam agents, amikacin (MIC50/90, 2/8μg/mL; 95.8/93.0% susceptible [CLSI/EUCAST]) was more active than colistin (MIC50/90, ≤0.5/>8μg/mL; 79.8% susceptible [EUCAST]), gentamicin (MIC50/90, ≤1/>8μg/mL; 72.0/71.3% susceptible [CLSI/EUCAST]), or levofloxacin (MIC50/90, 0.25/>4μg/mL; 65.7/63.8% susceptible [CLSI/EUCAST]). Against 495 ESBL non-CRE phenotype Enterobacteriaceae isolates, meropenem (MIC50/90, ≤0.06/0.12μg/mL; 97.0/100.0% susceptible [CLSI/EUCAST]), amikacin (MIC50/90, 4/16μg/mL; 95.1/89.7% susceptible), colistin (MIC50/90, ≤0.5/1μg/mL; 93.9% susceptible [EUCAST]), and ceftolozane-tazobactam (MIC50/90, 0.5/>32μg/mL; 74.7/66.9% susceptible) were the only agents to retain clinically useful activity (Table 2).
Activity of ceftolozane-tazobactam and comparator agents when tested against organisms and organism groups.
| Organism (no. tested) antimicrobial agent | MIC (μg/mL) | %S/%R | ||
|---|---|---|---|---|
| 50% | 90% | CLSIa | EUCASTa | |
| Enterobacteriaceae (1878) | ||||
| Ceftolozane-tazobactam | 0.25 | 32 | 84.2/14.1 | 80.9/19.1 |
| Amikacin | 2 | 8 | 95.8/2.9 | 93.0/4.2 |
| Cefepime | ≤0.5 | >16 | 65.2/29.5 | 63.4/32.9 |
| Ceftazidime | 0.25 | >16 | 66.8/28.8 | 61.8/33.2 |
| Colistin | ≤0.5 | >8 | 79.8/20.2 | |
| Gentamicin | ≤1 | >8 | 72.0/26.6 | 71.3/28.0 |
| Levofloxacin | 0.25 | >4 | 65.7/32.3 | 63.8/34.3 |
| Meropenem | ≤0.06 | 0.12 | 92.6/6.4 | 93.6/4.8 |
| Piperacillin-tazobactam | 2 | >64 | 80.2/13.8 | 75.7/19.8 |
| ESBL non-CRE Enterobacteriaceae (495) | ||||
| Ceftolozane-tazobactam | 0.5 | >32 | 74.7/21.4 | 66.9/33.1 |
| Amikacin | 4 | 16 | 95.1/3.2 | 89.7/4.9 |
| Cefepime | >16 | >16 | 9.7/78.3 | 6.5/86.4 |
| Ceftazidime | 16 | >16 | 19.2/67.3 | 6.5/80.8 |
| Colistin | ≤0.5 | 1 | 93.9/6.1 | |
| Gentamicin | >8 | >8 | 36.6/60.6 | 36.0/63.4 |
| Levofloxacin | >4 | >4 | 29.0/67.5 | 27.2/71.0 |
| Meropenem | ≤0.06 | 0.12 | 97.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 8 | >64 | 65.7/21.5 | 54.6/34.3 |
| Escherichia coli (661) | ||||
| Ceftolozane-tazobactam | 0.25 | 1 | 96.7/2.7 | 95.0/5.0 |
| Amikacin | 2 | 8 | 99.1/0.9 | 97.7/0.9 |
| Cefepime | ≤0.5 | >16 | 67.4/28.6 | 66.2/30.9 |
| Ceftazidime | 0.25 | >16 | 71.0/23.6 | 65.7/29.0 |
| Colistin | ≤0.5 | ≤0.5 | 99.8/0.2 | |
| Gentamicin | ≤1 | >8 | 71.6/27.6 | 71.2/28.4 |
| Levofloxacin | 0.5 | >4 | 54.2/43.6 | 53.2/45.8 |
| Meropenem | ≤0.06 | ≤0.06 | 99.7/0.3 | 99.7/0.0 |
| Piperacillin-tazobactam | 2 | 16 | 91.5/3.6 | 86.9/8.5 |
| ESBL non-CRE Escherichia coli (238) | ||||
| Ceftolozane-tazobactam | 0.5 | 2 | 91.6/6.7 | 87.0/13.0 |
| Amikacin | 4 | 8 | 97.5/2.5 | 94.9/2.5 |
| Cefepime | >16 | >16 | 11.0/78.9 | 7.6/85.2 |
| Ceftazidime | 16 | >16 | 20.2/64.7 | 5.5/79.8 |
| Colistin | ≤0.5 | ≤0.5 | 99.6/0.4 | |
| Gentamicin | >8 | >8 | 40.3/58.4 | 40.3/59.7 |
| Levofloxacin | >4 | >4 | 18.1/78.9 | 17.3/81.9 |
| Meropenem | ≤0.06 | ≤0.06 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 4 | 64 | 83.5/7.2 | 72.6/16.5 |
| Klebsiella pneumoniae (594) | ||||
| Ceftolozane-tazobactam | 0.5 | >32 | 64.6/33.3 | 60.4/39.6 |
| Amikacin | 2 | 16 | 90.6/6.6 | 86.9/9.4 |
| Cefepime | 8 | >16 | 45.5/48.4 | 44.4/52.6 |
| Ceftazidime | 8 | >16 | 47.5/46.8 | 43.6/52.5 |
| Colistin | ≤0.5 | 2 | 90.5/9.5 | |
| Gentamicin | ≤1 | >8 | 60.4/38.1 | 59.9/39.6 |
| Levofloxacin | 1 | >4 | 55.6/42.2 | 54.5/44.4 |
| Meropenem | ≤0.06 | >8 | 78.3/18.5 | 81.5/14.5 |
| Piperacillin-tazobactam | 8 | >64 | 58.6/34.0 | 52.9/41.4 |
| ESBL non-CRE Klebsiella pneumoniae (226) | ||||
| Ceftolozane-tazobactam | 2 | >32 | 56.6/38.5 | 46.0/54.0 |
| Amikacin | 2 | 16 | 93.4/3.5 | 86.7/6.6 |
| Cefepime | >16 | >16 | 6.7/80.4 | 4.0/90.2 |
| Ceftazidime | >16 | >16 | 11.5/76.5 | 2.2/88.5 |
| Colistin | ≤0.5 | 1 | 95.1/4.9 | |
| Gentamicin | >8 | >8 | 33.2/63.7 | 32.3/66.8 |
| Levofloxacin | >4 | >4 | 38.7/57.3 | 35.6/61.3 |
| Meropenem | ≤0.06 | 1 | 93.4/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 32 | >64 | 45.8/37.8 | 33.3/54.2 |
| Klebsiella oxytoca (54) | ||||
| Ceftolozane-tazobactam | 0.25 | 0.5 | 94.4/3.7 | 90.7/9.3 |
| Amikacin | 1 | 4 | 98.1/1.9 | 94.3/1.9 |
| Cefepime | ≤0.5 | 8 | 83.3/9.3 | 81.5/11.1 |
| Ceftazidime | ≤0.12 | 8 | 87.0/9.3 | 85.2/13.0 |
| Colistin | ≤0.5 | ≤0.5 | 98.1/1.9 | |
| Gentamicin | ≤1 | >8 | 85.2/14.8 | 83.3/14.8 |
| Levofloxacin | ≤0.12 | 0.5 | 94.4/3.7 | 90.7/5.6 |
| Meropenem | ≤0.06 | ≤0.06 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 2 | 64 | 88.7/7.5 | 86.8/11.3 |
| ESBL-phenotype Klebsiella oxytoca (11) | ||||
| Ceftolozane-tazobactam | 0.5 | 16 | 72.7/18.2 | 54.5/45.5 |
| Amikacin | 4 | 16 | 90.9/9.1 | 81.8/9.1 |
| Cefepime | 8 | >16 | 18.2/45.5 | 9.1/54.5 |
| Ceftazidime | 8 | >16 | 36.4/45.5 | 27.3/63.6 |
| Colistin | ≤0.5 | ≤0.5 | 90.0/10.0 | |
| Gentamicin | >8 | >8 | 36.4/63.6 | 36.4/63.6 |
| Levofloxacin | 0.5 | >4 | 81.8/18.2 | 81.8/18.2 |
| Meropenem | ≤0.06 | ≤0.06 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 64 | >64 | 45.5/36.4 | 36.4/54.5 |
| Enterobacter spp. (250) | ||||
| Ceftolozane-tazobactam | 0.5 | 8 | 84.0/14.0 | 79.2/20.8 |
| Amikacin | 1 | 8 | 96.8/2.0 | 94.0/3.2 |
| Cefepime | ≤0.5 | >16 | 72.3/20.1 | 68.3/24.1 |
| Ceftazidime | 0.5 | >16 | 66.0/31.2 | 59.6/34.0 |
| Colistin | ≤0.5 | >8 | 81.6/18.4 | |
| Gentamicin | ≤1 | >8 | 81.6/18.0 | 81.2/18.4 |
| Levofloxacin | ≤0.12 | >4 | 87.2/12.0 | 83.6/12.8 |
| Meropenem | ≤0.06 | ≤0.06 | 98.0/2.0 | 98.0/0.8 |
| Piperacillin-tazobactam | 4 | >64 | 79.0/10.5 | 75.8/21.0 |
| Ceftazidime-nonsusceptible (>4μg/mL) Enterobacter spp. (85) | ||||
| Ceftolozane-tazobactam | 2 | >32 | 52.9/41.2 | 38.8/61.2 |
| Amikacin | 2 | 16 | 90.6/5.9 | 85.9/9.4 |
| Cefepime | 16 | >16 | 31.0/54.8 | 19.0/61.9 |
| Ceftazidime | >16 | >16 | 0.0/91.8 | 0.0/100.0 |
| Colistin | ≤0.5 | >8 | 84.5/15.5 | |
| Gentamicin | ≤1 | >8 | 50.6/49.4 | 50.6/49.4 |
| Levofloxacin | 0.5 | >4 | 68.2/30.6 | 63.5/31.8 |
| Meropenem | ≤0.06 | 0.5 | 94.1/5.9 | 94.1/2.4 |
| Piperacillin-tazobactam | 32 | >64 | 44.0/27.4 | 35.7/56.0 |
| Citrobacter spp. (33) | ||||
| Ceftolozane-tazobactam | 0.25 | 8 | 69.7/18.2 | 63.6/36.4 |
| Amikacin | 1 | 2 | 93.9/6.1 | 93.9/6.1 |
| Cefepime | ≤0.5 | 2 | 93.9/3.0 | 87.9/6.1 |
| Ceftazidime | 0.5 | >16 | 69.7/30.3 | 63.6/30.3 |
| Colistin | ≤0.5 | 1 | 90.9/9.1 | |
| Gentamicin | ≤1 | ≤1 | 97.0/3.0 | 97.0/3.0 |
| Levofloxacin | ≤0.12 | 0.25 | 97.0/3.0 | 97.0/3.0 |
| Meropenem | ≤0.06 | ≤0.06 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 4 | 16 | 90.9/0.0 | 72.7/9.1 |
| Serratia marcescens (107) | ||||
| Ceftolozane-tazobactam | 0.5 | 1 | 95.3/2.8 | 91.6/8.4 |
| Amikacin | 2 | 4 | 99.1/0.9 | 92.5/0.9 |
| Cefepime | ≤0.5 | 2 | 91.6/4.7 | 86.9/8.4 |
| Ceftazidime | 0.25 | 1 | 92.5/5.6 | 90.7/7.5 |
| Colistin | >8 | >8 | 1.9/98.1 | |
| Gentamicin | ≤1 | 2 | 92.5/2.8 | 92.5/7.5 |
| Levofloxacin | ≤0.12 | 1 | 96.3/3.7 | 92.5/3.7 |
| Meropenem | ≤0.06 | ≤0.06 | 98.1/1.9 | 98.1/1.9 |
| Piperacillin-tazobactam | 2 | 8 | 97.2/2.8 | 93.5/2.8 |
| Proteus mirabilis (100) | ||||
| Ceftolozane-tazobactam | 0.5 | 1 | 98.0/1.0 | 96.0/4.0 |
| Amikacin | 2 | 8 | 98.0/1.0 | 94.0/2.0 |
| Cefepime | ≤0.5 | 16 | 85.0/12.0 | 85.0/15.0 |
| Ceftazidime | ≤0.12 | 0.5 | 97.0/1.0 | 93.0/3.0 |
| Colistin | >8 | >8 | 0.0/100.0 | |
| Gentamicin | ≤1 | >8 | 75.0/22.0 | 73.0/25.0 |
| Levofloxacin | ≤0.12 | >4 | 75.0/23.0 | 72.0/25.0 |
| Meropenem | ≤0.06 | 0.12 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | ≤0.5 | 1 | 98.0/0.0 | 97.0/2.0 |
| ESBL-phenotype Proteus mirabilis (17) | ||||
| Ceftolozane-tazobactam | 1 | 4 | 88.2/5.9 | 76.5/23.5 |
| Amikacin | 8 | 32 | 88.2/5.9 | 70.6/11.8 |
| Cefepime | 16 | >16 | 23.5/64.7 | 23.5/76.5 |
| Ceftazidime | 1 | 8 | 82.4/5.9 | 58.8/17.6 |
| Colistin | >8 | >8 | 0.0/100.0 | |
| Gentamicin | >8 | >8 | 29.4/58.8 | 23.5/70.6 |
| Levofloxacin | >4 | >4 | 11.8/88.2 | 11.8/88.2 |
| Meropenem | ≤0.06 | 0.12 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | 1 | 2 | 94.1/0.0 | 94.1/5.9 |
| Indole-positive Proteeae (60) | ||||
| Ceftolozane-tazobactam | 0.25 | 1 | 96.7/1.7 | 93.3/6.7 |
| Amikacin | 2 | 8 | 98.3/0.0 | 96.7/1.7 |
| Cefepime | ≤0.5 | 1 | 90.0/3.3 | 90.0/8.3 |
| Ceftazidime | ≤0.12 | 4 | 90.0/10.0 | 76.7/10.0 |
| Colistin | >8 | >8 | 0.0/100.0 | |
| Gentamicin | ≤1 | >8 | 81.7/16.7 | 76.7/18.3 |
| Levofloxacin | ≤0.12 | >4 | 83.3/11.7 | 76.7/16.7 |
| Meropenem | ≤0.06 | 0.12 | 100.0/0.0 | 100.0/0.0 |
| Piperacillin-tazobactam | ≤0.5 | 1 | 98.3/0.0 | 98.3/1.7 |
| Pseudomonas aeruginosa (537) | ||||
| Ceftolozane-tazobactam | 0.5 | 16 | 86.8/10.4 | 86.8/13.2 |
| Amikacin | 4 | 32 | 85.8/9.9 | 83.2/14.2 |
| Cefepime | 4 | >16 | 73.4/16.0 | 73.4/26.6 |
| Ceftazidime | 4 | >16 | 70.0/23.6 | 70.0/30.0 |
| Colistin | 1 | 2 | 98.9/0.0 | 100.0/0.0 |
| Gentamicin | 2 | >8 | 78.6/20.5 | 78.6/21.4 |
| Levofloxacin | 1 | >4 | 65.7/28.5 | 55.1/34.3 |
| Meropenem | 1 | >8 | 64.2/28.3 | 64.2/18.8 |
| Piperacillin-tazobactam | 8 | >64 | 68.9/16.4 | 68.9/31.1 |
| Ceftazidime-nonsusceptible (>8μg/mL) Pseudomonas aeruginosa (161) | ||||
| Ceftolozane-tazobactam | 4 | >32 | 56.5/34.8 | 56.5/43.5 |
| Amikacin | 8 | >32 | 66.5/24.2 | 61.5/33.5 |
| Cefepime | >16 | >16 | 23.6/50.9 | 23.6/76.4 |
| Ceftazidime | >16 | >16 | 0.0/78.9 | 0.0/100.0 |
| Colistin | 1 | 2 | 98.8/0.0 | 100.0/0.0 |
| Gentamicin | 8 | >8 | 49.1/49.7 | 49.1/50.9 |
| Levofloxacin | >4 | >4 | 28.6/63.4 | 20.5/71.4 |
| Meropenem | 8 | >8 | 24.8/61.5 | 24.8/48.4 |
| Piperacillin-tazobactam | >64 | >64 | 16.1/53.4 | 16.1/83.9 |
| Meropenem-nonsusceptible (>2μg/mL) Pseudomonas aeruginosa (192) | ||||
| Ceftolozane-tazobactam | 2 | >32 | 66.1/27.6 | 66.1/33.9 |
| Amikacin | 8 | >32 | 69.3/22.4 | 64.1/30.7 |
| Cefepime | 16 | >16 | 40.1/39.6 | 40.1/59.9 |
| Ceftazidime | >16 | >16 | 37.0/50.5 | 37.0/63.0 |
| Colistin | 1 | 2 | 99.0/0.0 | 100.0/0.0 |
| Gentamicin | 4 | >8 | 53.6/44.3 | 53.6/46.4 |
| Levofloxacin | >4 | >4 | 34.9/56.2 | 16.1/65.1 |
| Meropenem | >8 | >8 | 0.0/79.2 | 0.0/52.6 |
| Piperacillin-tazobactam | 64 | >64 | 34.4/35.4 | 34.4/65.6 |
A total of 661 E. coli isolates were evaluated, 96.7/95.0% of which were susceptible to ceftolozane-tazobactam (MIC50/90, 0.25/1μg/mL) by CLSI/EUCAST interpretive guidelines. Meropenem (MIC50/90, ≤0.06/≤0.06μg/mL; 99.7/99.7% susceptible [CLSI/EUCAST]), colistin (MIC50/90, ≤0.5/≤0.5μg/mL; 99.8% susceptible [EUCAST]), amikacin (MIC50/90, 2/8μg/mL; 99.1/97.7% susceptible), and piperacillin-tazobactam (MIC50/90, 2/16μg/mL; 91.5/86.9% susceptible) showed good activity against E. coli, followed by cefepime (MIC50/90, ≤0.5/>16μg/mL; 67.4/66.2% susceptible), ceftazidime (MIC50/90, 0.25/>16μg/mL; 71.0/65.7% susceptible), and gentamicin (MIC50/90, ≤1/>8μg/mL; 71.6/71.2% susceptible) (Table 2). Among ESBL non-CRE phenotype isolates of E. coli, resistance rates to cefepime, ceftazidime, gentamicin, and levofloxacin were elevated (Table 2). Meropenem (MIC50/90, ≤0.06/≤0.06μg/mL; 100.0/100.0% susceptible) and amikacin (MIC50/90, 4/8μg/mL; 97.5/94.9% susceptible) retained potent activity against ESBL non-CRE phenotype strains of E. coli. For ESBL non-CRE phenotype isolates, ceftolozane-tazobactam inhibited 91.6% at ≤2μg/mL and 93.3% at ≤4μg/mL (Tables 1 and 2). Piperacillin-tazobactam showed moderate activity (MIC50/90, 4/64μg/mL; 83.5/72.6% susceptible) against these E. coli strains.
Ceftolozane-tazobactam showed potent activity against non-ESBL phenotype isolates of K. pneumoniae (MIC50/90, 0.25/0.5μg/mL; highest MIC, 2μg/mL) and retained activity against many ESBL non-CRE phenotype isolates (MIC50/90, 2/>32μg/mL; 56.6/46.0% susceptible) (Tables 1 and 2). Among the β-lactam comparator agents tested, only meropenem was more active than ceftolozane-tazobactam against Klebsiella species irrespective of the resistant phenotype (Table 2). Ceftolozane-tazobactam was also active against other frequently isolated Enterobacteriaceae, including K. oxytoca (MIC50/90, 0.25/0.5μg/mL), Enterobacter spp. (MIC50/90, 0.5/8μg/mL), Citrobacter spp. (MIC50/90, 0.25/8μg/mL), P. mirabilis (MIC50/90, 0.5/1μg/mL), indole-positive Proteeae (MIC50/90, 0.25/1μg/mL), and Serratia marcescens (MIC50/90, 0.5/1μg/mL) (Tables 1 and 2).
Activities of ceftolozane-tazobactam and comparators against P. aeruginosaCeftolozane-tazobactam was the most potent (MIC50//90, 0.5/16μg/mL) β-lactam agent tested against 537 P. aeruginosa isolates, inhibiting 86.8% at an MIC of ≤4μg/mL (Tables 1 and 2). Based on the MIC50 value, ceftolozane-tazobactam was 2-fold more active than meropenem (MIC50, 1μg/mL), 8-fold more active than cefepime (MIC50, 4μg/mL) and ceftazidime (MIC50, 4μg/mL), and 16-fold more active than piperacillin-tazobactam (MIC50, 8μg/mL) (Table 2). Overall susceptibility rates (by CLSI/EUCAST criteria; Table 2) for cefepime (73.4/73.4%), ceftazidime (70.0/70.0%), meropenem (64.2/64.2%), and piperacillin-tazobactam (68.9/68.9%) were all below that of ceftolozane-tazobactam at ≤4μg/mL (86.8/86.8%; Table 2). Amikacin (MIC50/90, 4/32μg/mL; 85.8/83.2% susceptible) and colistin (MIC50/90, 1/2μg/mL; 98.9/100.0% susceptible) showed good activity against P. aeruginosa (Table 2).
Ceftolozane-tazobactam retained moderate activity against P. aeruginosa isolates that were NS to ceftazidime (56.5% susceptible) and meropenem (66.1% susceptible), the other antipseudomonal β-lactam agents (Table 2). No other β-lactam agents inhibited more than 40.1% of these resistant phenotypes. Notably, colistin was highly active against both ceftazidime-NS (MIC50/90, 1/2μg/mL; 98.8/100.0% susceptible) and meropenem-NS (MIC50/90, 1/2μg/mL; 99.0/100.0% susceptible) strains of P. aeruginosa (Table 2).
DiscussionThe results of the present study extend those previously reported concerning the in vitro activity of ceftolozane-tazobactam against European strains of Enterobacteriaceae and P. aeruginosa.13–15 Although the rates of β-lactam resistance among isolates from Latin America were much higher than seen in Europe, ceftolozane-tazobactam was the most active of the tested β-lactam agents against P. aeruginosa and was second to meropenem against Enterobacteriaceae. Whereas ceftolozane-tazobactam had little activity against CRE strains of Enterobacteriaceae, it retained activity against most ESBL non-CRE phenotype strains, second only to meropenem. Likewise, ceftolozane-tazobactam was more active than the other antipseudomonal β-lactam agents tested against strains of P. aeruginosa that were NS to ceftazidime and meropenem. Among the non-β-lactam comparator agents, colistin and amikacin were the most active against Enterobacteriaceae and P. aeruginosa, including the various resistant phenotypes.
In agreement with previous surveys,2,3 we found great variation in Enterobacteriaceae and P. aeruginosa resistance rates to ceftazidime and meropenem among Latin American countries (Tables 3 and 4). This variation has been shown to correlate with regional differences in β-lactamase production.14 Previously (2008–2011), we have shown that among Latin American isolates of E. coli and K. pneumoniae, the rates of ESBL and CRE phenotypes varied markedly from country to country.2,3 As seen in Table 3, resistance rates among these two species from 2013 to 2015 show continued variability among the Latin American nations evaluated. The occurrence of K. pneumoniae isolates with a CRE phenotype ranged from 1.9% in Chile to 33.9% in Brazil and was 19.2% overall. The highest rates of ESBL non-CRE phenotype E. coli and K. pneumoniae were found in isolates from Chile (29.8% and 67.0%, respectively) and Mexico (66.8% and 44.3%) and the lowest rates were in isolates from Brazil (13.0% and 21.6%) and Argentina (16.2% and 34.8%). These data are comparable to those reported in 2011 for E. coli; however, the frequency of CRE strains of K. pneumoniae in the present survey (19.2%; Table 3) is considerably higher than that reported in 2011 (9.0%).3
Geographical distribution of phenotypically resistant isolates.
| Country | % of isolates with resistant phenotypes (no. resistant/no. tested) | ||||||
|---|---|---|---|---|---|---|---|
| CRE | ESBL non-CRE | CAZ-NS | MER-NS | ||||
| KPN | EC | KPN | ENTB | PSA | KPN | PSA | |
| Argentina | 18.3% (21/115) | 16.2% (16/99) | 34.8% (40/115) | 32.4% (11/34) | 38.7% (46/119) | 21.7% (25/115) | 35.3% (42/119) |
| Brazil | 33.9% (74/218) | 13.0% (26/200) | 21.6% (47/218) | 34.7% (26/75) | 23.0% (49/213) | 35.3% (77/218) | 35.7% (76/213) |
| Chile | 1.9% (2/103) | 29.8% (37/124) | 67.0% (69/103) | 28.9% (13/45) | 50.6% (45/89) | 9.7% (10/103) | 48.3% (43/89) |
| Mexico | 10.8% (17/158) | 66.8% (159/238) | 44.3% (70/158) | 36.5% (35/96) | 18.1% (21/116) | 10.8% (17/158) | 26.7% (31/116) |
| Total | 19.2% (114/594) | 36.0% (238/661) | 38.0% (226/594) | 34.0% (85/250) | 30.0% (161/537) | 21.7% (129/594) | 35.8% (192/537) |
CRE, carbapenem resistant Enterobacteriaceae; ESBL, extended-spectrum β-lactamase; CAZ-NS, ceftazidime-nonsusceptible; MER-NS, meropenem-nonsusceptible; KPN, Klebsiella pneumoniae; EC, Escherichia coli; ENTB, Enterobacter spp.; PSA, Pseudomonas aeruginosa.
Antimicrobial activity of ceftolozane-tazobactam, ceftazidime, and meropenem against Latin American strains of P. aeruginosa stratified by country.
| Country (no. tested) | % susceptible (no. susceptible)a | ||
|---|---|---|---|
| Ceftolozane-tazobactam | Ceftazidime | Meropenem | |
| Argentina (119) | 83.2 (99) | 61.3 (73) | 64.7 (77) |
| Brazil (213) | 90.6 (193) | 77.0 (164) | 64.3 (137) |
| Chile (89) | 77.5 (69) | 49.4 (44) | 51.7 (46) |
| Mexico (116) | 90.5 (105) | 81.9 (95) | 73.3 (85) |
| Total (537) | 86.8 (466) | 70.0 (376) | 64.2 (345) |
As observed in 2008–2010,14P. aeruginosa susceptibility to antipseudomonal β-lactams and the frequency of resistant phenotypes varied markedly among the Latin American nations that participated in the survey (Tables 3 and 4). The lowest susceptibility rates for ceftazidime and meropenem were observed in Chile (49.4% and 51.7%, respectively) and the highest in Mexico (81.9% and 73.3%). Ceftolozane-tazobactam activity was also compromised when tested against P. aeruginosa isolates from Chile (77.5% susceptible), but it provided greater than 90% coverage against isolates from Brazil and Mexico (Table 4).
Whereas the spread of CRE is a major concern worldwide, the prevalence is variable with considerably higher rates in Latin America than in Europe or North America.14,20 Our results confirm these observations with CRE rates exceeding 10% in Argentina (18.3%), Brazil (33.9%), and Mexico (10.8%) (Table 3). High and variable rates of ESBL non-CRE phenotype and MDR Enterobacteriaceae are consistent with those previously reported by Farrell et al.,21 Gales et al.,2 Jones et al.,3 Sader et al.,14 and Canton et al.20
ConclusionsIn conclusion, these data for ceftolozane-tazobactam collected from 2013 through 2015 from 12 medical centers in four Latin American countries demonstrated sustained potency and spectrum against Enterobacteriaceae and P. aeruginosa in agreement with previous studies.14,15 These data suggest that ceftolozane-tazobactam may be an important treatment option for HAIs caused by wild-type and MDR strains of P. aeruginosa and Enterobacteriaceae.10 One of the more important aspects of this survey is the confirmation of extensive variation in antibiotic-resistant phenotypes of GNB in Latin America. This finding emphasizes the need for ongoing surveillance and application of antimicrobial stewardship to prevent the further spread of β-lactam resistance.9
Conflicts of interestJMI Laboratories contracted to perform services in 2016 for Achaogen, Actelion, Allecra Therapeutics, Allergan, AmpliPhi Biosciences, API, Astellas Pharma, AstraZeneca, Basilea Pharmaceutica, Bayer AG, BD, Biomodels, Cardeas Pharma Corp., CEM-102 Pharma, Cempra, Cidara Therapeutics, Inc., CorMedix, CSA Biotech, Cutanea Life Sciences, Inc., Debiopharm Group, Dipexium Pharmaceuticals, Inc., Duke, Entasis Therapeutics, Inc., Fortress Biotech, Fox Chase Chemical Diversity Center, Inc., Geom Therapeutics, Inc., GSK, Laboratory Specialists, Inc., Medpace, Melinta Therapeutics, Inc., Merck & Co., Inc., Micromyx, MicuRx Pharmaceuticals, Inc., Motif Bio, N8 Medical, Inc., Nabriva Therapeutics, Inc., Nexcida Therapeutics, Inc., Novartis, Paratek Pharmaceuticals, Inc., Pfizer, Polyphor, Rempex, Scynexis, Shionogi, Spero Therapeutics, Symbal Therapeutics, Synlogic, TenNor Therapeutics, TGV Therapeutics, The Medicines Company, Theravance Biopharma, ThermoFisher Scientific, VenatoRx Pharmaceuticals, Inc., Wockhardt, Zavante Therapeutics, Inc. There are no speakers’ bureaus or stock options to declare.
Funding for this research was provided by Merck & Co., Inc., Kenilworth, NJ, USA.