The mechanisms involved in the uncommon resistance phenotype, carbapenem resistance and broad-spectrum cephalosporin susceptibility, were investigated in 25 Pseudomonas aeruginosa clinical isolates that exhibited this phenotype, which were recovered from three different hospitals located in São Paulo, Brazil. The antimicrobial susceptibility profile was determined by CLSI broth microdilution. β-lactamase-encoding genes were investigated by PCR followed by DNA sequencing. Carbapenem hydrolysis activity was investigated by spectrophotometer and MALDI-TOF assays. The mRNA transcription level of oprD was assessed by qRT-PCR and the outer membrane proteins profile was evaluated by SDS-PAGE. Genetic relationship among P. aeruginosa isolates was assessed by PFGE. Carbapenems hydrolysis was not detected by carbapenemase assay in the carbapenem-resistant and cephalosporin-susceptible P. aueruginosa clinical isolates. OprD decreased expression was observed in all P. aeruginosa isolates by qRT-PCR. The outer membrane protein profile by SDS-PAGE suggested a change in the expression of the 46kDa porin that could correspond to OprD porin. The isolates were clustered into 17 genotypes without predominance of a specific PFGE pattern. These results emphasize the involvement of multiple chromosomal mechanisms in carbapenem-resistance among clinical isolates of P. aeruginosa, alert for adaptation of P. aeruginosa clinical isolates under antimicrobial selective pressure and make aware of the emergence of an uncommon phenotype among P. aeruginosa clinical isolates.
Pseudomonas aeruginosa is one of the most frequent pathogens associated to nosocomial infections, especially among immunocompromised patients1 and exhibits notorious versatility and capacity to acquire resistance mechanisms to antimicrobial therapy.2 Beta-lactam antimicrobial agents are the main option to treat serious infection caused by this pathogen. However, the production of β-lactamases, such as cephalosporinases and carbapenemases, has been intensely reported among clinical isolates of P. aeruginosa from Latin America and represents the most effective mechanism of β-lactams resistance reported among Gram-negative worldwide.3
Since the carbapenems molecules are more resistant to hydrolysis activity by a great number of spread serine-β-lactamases, these drugs have a particular value in the treatment of infections caused by cephalosporinases producer strains,4 which remain susceptible to carbapenems. In P. aeruginosa, the carbapenems resistance is modulated by acquired carbapenemases in association with intrinsic mechanisms such as down-regulation or loss of OprD porin, efflux pumps hyperexpression, chromosomal AmpC β-lactamase production, and target alterations.2,3 However, since carbapenemases have the ability to hydrolyze penicillins, cephalosporins, besides carbapenems, Gram-negative bacteria carrying a carbapenemase-encoding gene frequently exhibit resistance to virtually all β-lactams.5
Given the importance of carbapenem for the treatment of infections caused by P. aeruginosa, it is essential to clarify the mechanisms involved in unusual and/or poorly known phenotypes. Knowledge of these mechanisms alert for an adaptation to the selective pressure exerted by antimicrobial and drug resistance development, thus affecting the treatment of infections caused by these pathogens often restricted to only polymyxins.
The aim of this study was to analyze the possible mechanism of antimicrobial resistance involved in clinical isolates of P. aeruginosa that exhibited an uncommon phenotype of resistance: resistance to carbapenems but susceptibility to broad-spectrum cephalosporins (Carb-R/Ceph-S).
MethodsBacterial isolates, identification and antimicrobial susceptibility testingBetween May, 2012 and March, 2013, a total of 25 P. aeruginosa clinical isolates exhibiting carbapenem resistance but broad-spectrum cephalosporin susceptibility (Carb-R/Ceph-S) were recovered from 18 different infected patients from three distinct hospitals located in São Paulo, Brazil (Table 1). The identification at the species level was confirmed by MALDI-TOF MS in a Bruker Daltonics Microflex LT MALDI-TOF using the Biotyper MALDI 2.0 (Bruker Daltonics, Bremen, Germany) as previously described.6 The susceptibility profile was confirmed by broth microdilution to imipenem, meropenem, ceftazidime, cefepime, amikacin, gentamicin, and ciprofloxacin according the CLSI recommendations.7,8 American Type Culture Collection (ATCC) Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 29213 strains were used as susceptibility testing quality control.
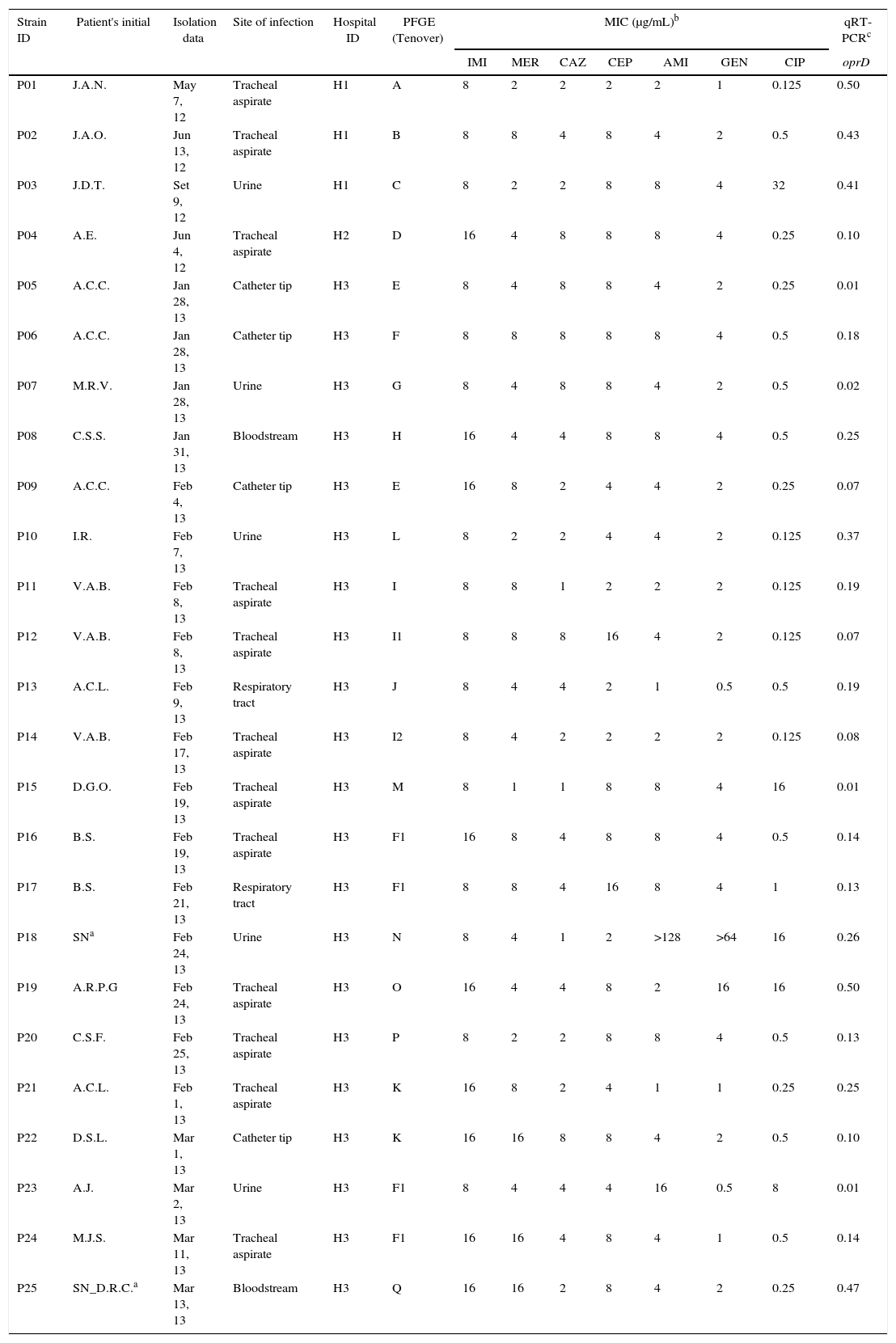
Carb-R/Ceph-S P. aeruginosa clinical isolates according to the hospital, susceptibility profile, and oprD relative transcriptional level.
| Strain ID | Patient's initial | Isolation data | Site of infection | Hospital ID | PFGE (Tenover) | MIC (μg/mL)b | qRT-PCRc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMI | MER | CAZ | CEP | AMI | GEN | CIP | oprD | ||||||
| P01 | J.A.N. | May 7, 12 | Tracheal aspirate | H1 | A | 8 | 2 | 2 | 2 | 2 | 1 | 0.125 | 0.50 |
| P02 | J.A.O. | Jun 13, 12 | Tracheal aspirate | H1 | B | 8 | 8 | 4 | 8 | 4 | 2 | 0.5 | 0.43 |
| P03 | J.D.T. | Set 9, 12 | Urine | H1 | C | 8 | 2 | 2 | 8 | 8 | 4 | 32 | 0.41 |
| P04 | A.E. | Jun 4, 12 | Tracheal aspirate | H2 | D | 16 | 4 | 8 | 8 | 8 | 4 | 0.25 | 0.10 |
| P05 | A.C.C. | Jan 28, 13 | Catheter tip | H3 | E | 8 | 4 | 8 | 8 | 4 | 2 | 0.25 | 0.01 |
| P06 | A.C.C. | Jan 28, 13 | Catheter tip | H3 | F | 8 | 8 | 8 | 8 | 8 | 4 | 0.5 | 0.18 |
| P07 | M.R.V. | Jan 28, 13 | Urine | H3 | G | 8 | 4 | 8 | 8 | 4 | 2 | 0.5 | 0.02 |
| P08 | C.S.S. | Jan 31, 13 | Bloodstream | H3 | H | 16 | 4 | 4 | 8 | 8 | 4 | 0.5 | 0.25 |
| P09 | A.C.C. | Feb 4, 13 | Catheter tip | H3 | E | 16 | 8 | 2 | 4 | 4 | 2 | 0.25 | 0.07 |
| P10 | I.R. | Feb 7, 13 | Urine | H3 | L | 8 | 2 | 2 | 4 | 4 | 2 | 0.125 | 0.37 |
| P11 | V.A.B. | Feb 8, 13 | Tracheal aspirate | H3 | I | 8 | 8 | 1 | 2 | 2 | 2 | 0.125 | 0.19 |
| P12 | V.A.B. | Feb 8, 13 | Tracheal aspirate | H3 | I1 | 8 | 8 | 8 | 16 | 4 | 2 | 0.125 | 0.07 |
| P13 | A.C.L. | Feb 9, 13 | Respiratory tract | H3 | J | 8 | 4 | 4 | 2 | 1 | 0.5 | 0.5 | 0.19 |
| P14 | V.A.B. | Feb 17, 13 | Tracheal aspirate | H3 | I2 | 8 | 4 | 2 | 2 | 2 | 2 | 0.125 | 0.08 |
| P15 | D.G.O. | Feb 19, 13 | Tracheal aspirate | H3 | M | 8 | 1 | 1 | 8 | 8 | 4 | 16 | 0.01 |
| P16 | B.S. | Feb 19, 13 | Tracheal aspirate | H3 | F1 | 16 | 8 | 4 | 8 | 8 | 4 | 0.5 | 0.14 |
| P17 | B.S. | Feb 21, 13 | Respiratory tract | H3 | F1 | 8 | 8 | 4 | 16 | 8 | 4 | 1 | 0.13 |
| P18 | SNa | Feb 24, 13 | Urine | H3 | N | 8 | 4 | 1 | 2 | >128 | >64 | 16 | 0.26 |
| P19 | A.R.P.G | Feb 24, 13 | Tracheal aspirate | H3 | O | 16 | 4 | 4 | 8 | 2 | 16 | 16 | 0.50 |
| P20 | C.S.F. | Feb 25, 13 | Tracheal aspirate | H3 | P | 8 | 2 | 2 | 8 | 8 | 4 | 0.5 | 0.13 |
| P21 | A.C.L. | Feb 1, 13 | Tracheal aspirate | H3 | K | 16 | 8 | 2 | 4 | 1 | 1 | 0.25 | 0.25 |
| P22 | D.S.L. | Mar 1, 13 | Catheter tip | H3 | K | 16 | 16 | 8 | 8 | 4 | 2 | 0.5 | 0.10 |
| P23 | A.J. | Mar 2, 13 | Urine | H3 | F1 | 8 | 4 | 4 | 4 | 16 | 0.5 | 8 | 0.01 |
| P24 | M.J.S. | Mar 11, 13 | Tracheal aspirate | H3 | F1 | 16 | 16 | 4 | 8 | 4 | 1 | 0.5 | 0.14 |
| P25 | SN_D.R.C.a | Mar 13, 13 | Bloodstream | H3 | Q | 16 | 16 | 2 | 8 | 4 | 2 | 0.25 | 0.47 |
Acquired β-lactamases encoding genes were investigated by PCR and DNA sequencing, as previously described, using primers for: cephalosporinases (blaAmpC); (b) serino-β-lactamases (blaTEM, blaSHV, blaGES, blaCTX-M, blaBES, blaPER, blaKPC, blaSME); (c) oxacillinases (blaOXA-1, blaOXA-2, blaOXA-3, blaOXA-5, blaOXA-7, blaOXA-18, blaOXA-45, blaOXA-46, blaOXA-50, blaOXA-23, blaOXA-24, blaOXA-51, blaOXA-58, blaOXA-20, blaOXA-48, blaOXA-62, blaOXA-143, blaOXA-198); (d) metallo-β-lactamases (blaIMP, blaVIM, blaSPM, blaGIM, blaSIM, blaNDM); and (e) class 1 integron.9 Amplicons were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced in both strands using the Applied Biosystems 3500 genetic analyzer equipment (Applied Biosystems, PerkinElmer, USA). The nucleotide sequence and the respective deduced amino acid sequences were analyzed using the Lasergene software package (DNAStar, Madison, WI, USA) and compared with the sequences available on the Internet using the BLAST tool (http://www.ncbi.nlm.nih.gov/blast/).
Carbapenemase hydrolysis assayCarbapenemase activity was investigated in bacterial cell crude extracts by UV spectrophotometric assays against 100μM imipenem and 100μM meropenem in 100mM phosphate buffer (pH 7.0), as previously described.10 In parallel, carbapenem hydrolysis inhibition was performed by incubating the whole-protein extract with 25mM EDTA for 15min, previously to the assay with imipenem and meropenem. Imipenem hydrolysis was also investigated by MALDI-TOF MS according to the method described by Carvalhaes and colleagues.11
Relative gene transcriptional levelRelative transcriptional levels of mexB, mexD, mexF, mexY, ampC, and oprD were determined and analyzed with Real Time 7500 (Applied Biosystems, Warrington, United Kingdom) as previously described.12 Quantitative RT-PCR was performed with Platinum SYBR Green Supermix (Invitrogen, Carlsbad, USA). According to previous studies, reduced oprD and increased ampC transcription levels were considered significant when it was ≤70% and ≥10-fold, respectively, compared to their transcriptional levels in PAO1 strain.13 MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY efflux systems were considered overexpressed when mexB, mexC, mexE, and mexY transcriptional level were at least 2-, 100-, 100-, and 4-fold higher than those in PAO1 strain, respectively.14
Assessment of the OMP profile and oprD analysisAmplification of oprD gene was carried out by conventional PCR as previously described15 and the amplicon size analyzed by 1.5% agarose gel electrophoresis. The outer membrane proteins (OMP) were analyzed by SDS-PAGE in a 15% acrylamide and 0.3% N,N′-methylenebisacrylamide running gel. The proteins were stained with coomassie blue r250 and photographed using GelDoc System Imaging. P. aeruginosa PA01 and ATCC 27853 were used as reference strains.16
Molecular typing by pulsed-field gel electrophoresisGenomic DNA of isolates was directly prepared from a bacterial cell suspension into agarose blocks and digested with SpeI restriction enzyme (New England, Beverly, MA, USA). Electrophoresis was performed on CHEF-DR III (BioRad, Richmond, CA). The band patterns were analyzed by BioNumerics 2.0 software (Applied Maths, Belgium)17,18 and according to Tenover interpretive criteria.19
ResultsCarbapenem-resistance not related to carbapenemaseThe decreased susceptibility to at least one tested carbapenem was confirmed as well as the susceptibility to tested cephalosporins as shown in Table 1. All evaluated isolates exhibited resistance to imipenem with MIC range varying from 8 to 16μg/mL. Although five isolates (20%) showed meropenem susceptibility (MIC≤2), most isolates had reduced susceptibility to this antimicrobial (MICs≥4μg/mL). All P. aeruginosa isolates were susceptible to ceftazidime (MIC≤8). Only two isolates exhibited a cefepime MIC equal to 16μg/mL, and were classified as intermediate resistant to cefepime according CLSI breakpoints.8
All 25 P. aeruginosa isolates carried the constitutive blaOXA-50 gene showing several DNA silent mutations but all of them presented the predicted STYK motif commonly found in class D β-lactamases (DBLs), which have narrow β-lactamase hydrolysis activity.20 A single isolate (P18) also had positive PCR for blaOXA-56 gene. DNA sequencing analysis of the blaOXA-56 genetic context showed that it was carried by a class 1 integron (named In163) as previously described by Carvalho et al. among SPM-1 producing P. aeruginosa clinical isolates.21 None tested carbapenemase encoding genes were detected by PCR.
To determine whether the carbapenem-resistance was mediated by an unknown carbapenemase, in vitro hydrolysis of imipenem and meropenem by whole-protein crude cell extract was investigated by spectrophotometric and MALDI-TOF MS analysis. None of the tested clinical sample were able to hydrolyze imipenem and meropenem molecules in a spectrophotometric assay (data not shown). Furthermore, no carbapenem hydrolysis was detected by MALDI-TOF MS.
Mechanism associated with carbapenem-resistanceThe expression of OprD was estimated by mRNA transcriptional level by qRT-PCR and outer membrane protein pattern by SDS-PAGE. Amplicons with size higher than expected for wild type P. aeruginosa were observed for P05, P09, P13, P15, and P21 isolates. The amplification of oprD gene failed in two isolates (P07 and P22). Relative transcriptional level was performed to evaluate oprD down-regulation. Relative transcriptional level analysis using P. aeruginosa PA01 as reference stain (baseline) showed that the transcriptional level of oprD gene among those isolates was significantly reduced when compared to wild-type PA01 strain.
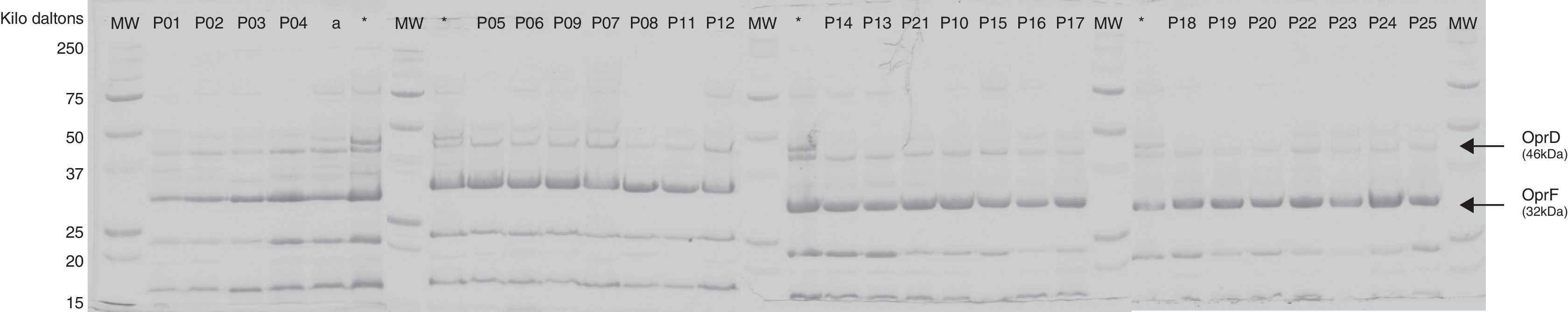
The evaluation of the OMP by SDS-PAGE strongly suggested that OprD was visually absent or produced at very low levels by all isolates (Fig. 1).
Outer membrane pattern of Carb-R/Ceph-S P. aeruginosa clinical isolates. Outer membrane profiles were analyzed for the presence of the porin, OprD. PAO1* and ATCC 27853a were included as controls. The OprD and OprF protein bands are indicated with arrow. MW, molecular weight protein maker (kDa).
Overall, the mRNA transcription levels of mexB, mexC, mexE, mexY and also of the β-lactamase AmpC of most Carb-R/Ceph-S P. aeruginosa isolates were even lower than those observed for PAO1 strain (data not shown).
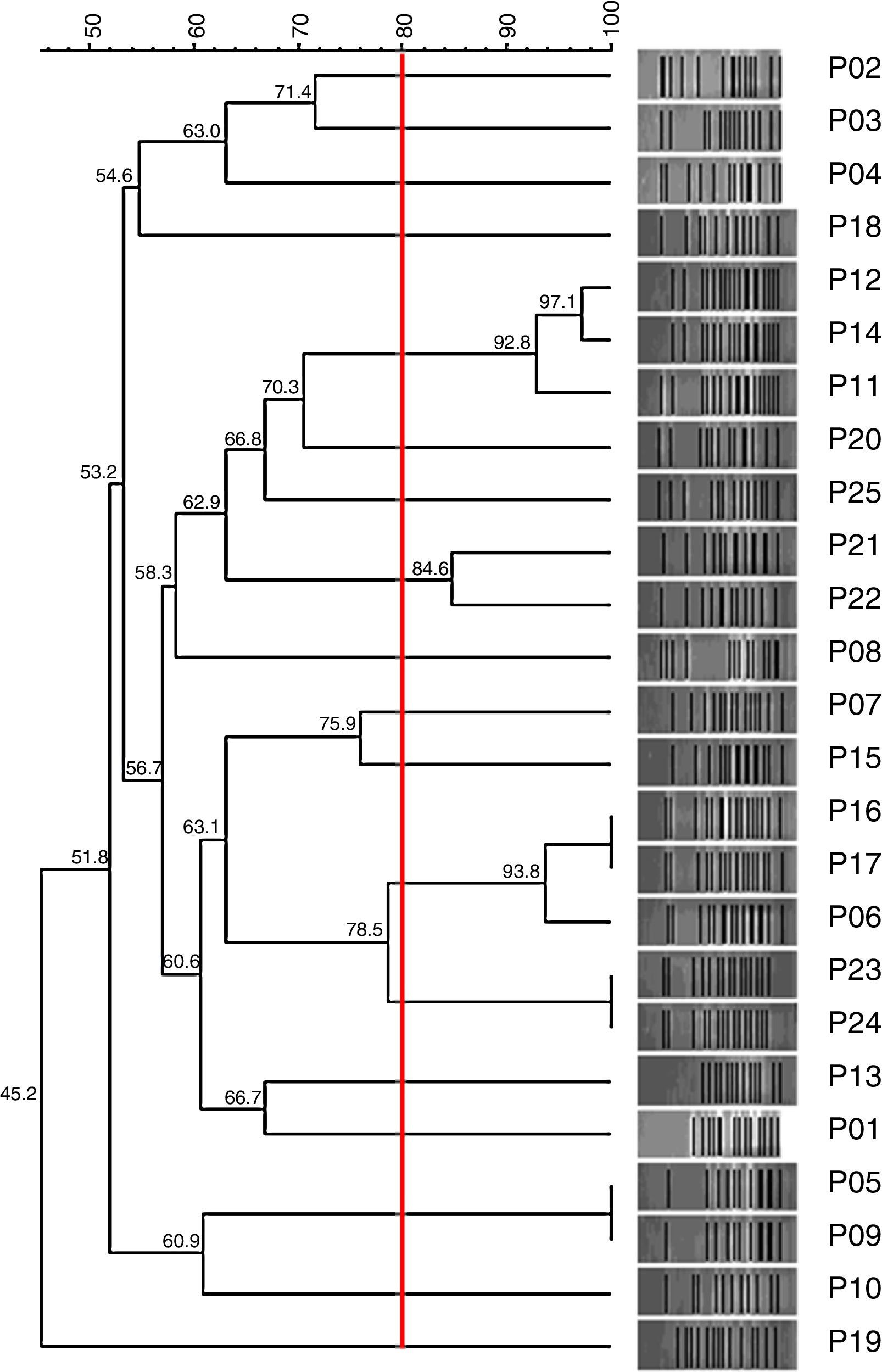
Genetic relationshipHigh clonal diversity was observed between the 25 tested clinical isolates of P. aeruginosa, with 17 different PFGE patterns without predominance of a single pattern (Fig. 2).
DiscussionCarbapenems have been extremely prescribed for treatment of P. aeruginosa infections.2 Carbapenemase production has emerged as the main mechanism of carbapenem resistance among clinical isolates of P. aeruginosa. However, loss of OprD has also contributed for carbapenem resistance, especially conferring resistance to imipenem. Acquisition of mutations, insertions, and/or deletions in the oprD gene are the most common mechanisms of OprD inactivation as well as down-regulation of oprD transcription.22
This study evaluated clinical isolates of P. aeruginosa exhibiting reduced susceptibility to carbapenems not related to carbapenemase production, which has been mostly related to carbapenemase in Brazilian clinical settings.12,23 The emergence of P. aeruginosa resistant to carbapenems associated with loss of porin has been documented in clinical settings and also could be selected in vitro after laboratory carbapenems exposure. In a study carried out by Fowler and colleagues, which evaluated the mechanism of carbapenems resistance in selected mutants after exposure of a cystic fibrosis clinical isolate to sub-inhibitory concentration of meropenem, demonstrated that loss of OprD by an IS element (ISPa8) disruption increased the MIC to imipenem and meropenem from 0.5 and 2μg/mL, respectively, to up to 16μg/mL.24 In another work conducted by Ocampo-Sosa and colleagues, OprD modifications were also found among carbapenem susceptible isolates, with imipenem MICs varying from 0.06 to 4μg/mL in a clinical setting of metallo-β-lactamase-negative P. aeruginosa isolated from Spanish hospitals.16
In the present study we analyzed P. aeruginosa isolates resistant to at least one carbapenem with imipenem MICs ranging from 8 to 16μg/mL and meropenem MIC varying from 1 to 16μg/mL that remained susceptible to broad-spectrum cephalosporins. All isolates analyzed had a significant reduced expression or lack of OprD porin in P. aeruginosa outer membrane by expression of a null or altered oprD gene.
We found a high diversity of genotypes among the OprD-deficient P. aeruginosa, which were isolated from three different hospitals located in São Paulo city, Brazil. It may reveal an emergence of OprD-deficient P. aeruginosa in the Brazilian clinical scenario by a manner independently of a clone spread outwards, in accordance with previous observation.25 The ability of OprD-mutant selection by carbapenems exposure may play an important role to OprD-mutant emergence and it should be considered regarding empirical clinical usage of carbapenems.
Loss of OprD has also been implicated in an increased in vivo fitness and/or virulence in a murine infection model. OprD-deficient P. aeruginosa strains were more resistant to low pH environment, to normal human serum, and presented an increased cytotoxicity against murine macrophages. It has suggested that the association of antimicrobial resistance and increased survival of these strains can favor the permanence of this strains in the hospital setting and make treatment more difficult.26 The increase in OprD-deficient survival can also raise the persistence of these strains in the hospital environment and favor the acquisition of new determinant of antimicrobial resistance by horizontal gene transfer.27 It is worth mentioning that all isolates evaluated in this study were susceptible to broad-spectrum cephalosporins (ceftazidime and cefepime). As previously demonstrated, OprD mutants are resistant only to zwitterionic carbapenems, especially imipenem and the cross-resistance to others β-lactams has not been observed in these strains. Carb-R/Ceph-S phenotype might be used as a phenotypic biomarker for screening OprD-deficient P. aeruginosa clinical isolates. Since these P. aeruginosa do not produce high levels of AmpC, theoretically ceftazidime or cefepime could be prescribed for treatment of such infections. However, more studies would be necessary to ensure the efficacy of broad-spectrum cephalosporins for treatment of Carb-R/Ceph-S P. aeruginosa infections.
ConclusionThe carbapenem resistance of P. aeruginosa associated to OprD-deficient may become more frequent in the hospitals scenario as consequence of selective pressure exerted by clinical use of carbapenems. Considering the therapeutic value of carbapenems as one of the last options for the treatment of P. aeruginosa infectious and the emergence of P. aeruginosa as a public health problem, rational carbapenems usage is essential to reduce the selective pressure over P. aeruginosa clinical isolates.
FundingThis study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant number: 2010/12891-9) and Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Conflicts of interestThe authors declare no conflicts of interest.
We would like to thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Ministry of Science and Technology (Brazil), for providing a research grant to A.C.G. (305535/2014-5). We also would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and to CNPq for granting the Doctoral scholar fellowship to E.H.C. and to D.E.X. (CNPq 141572/2008-6).