Biofilm production is an important mechanism for the survival of Pseudomonas aeruginosa and its relationship with antimicrobial resistance represents a challenge for patient therapeutics. P. aeruginosa is an opportunistic pathogen frequently associated to nosocomial infections, especially in imunocompromised hosts.
ObjectivesAnalyze the phenotypic biofilm production in P. aeruginosa isolates, describe clonal profiles, and analyze quorum sensing (QS) genes and the occurrence of mutations in the LasR protein of non-biofilm producing isolates.
MethodsIsolates were tested for biofilm production by measuring cells adherence to the microtiter plates. Clonal profile analysis was carried out through ERIC-PCR, QS genes were by specific PCR.
ResultsThe results showed that 77.5% of the isolates were considered biofilm producers. The results of genotyping showed 38 distinct genetic profiles. As for the occurrence of the genes, 100% of the isolates presented the lasR, rhlI and rhlR genes, and 97.5%, presented the lasI gene. In this study nine isolates were not biofilm producers. However, all presented the QS genes. Amplicons related to genes were sequenced in three of the nine non-biofilm-producing isolates (all presenting different genetic similarity profile) and aligned to the sequences of those genes in P. aeruginosa strain PAO1 (standard biofilm-producing strain). Alignment analysis showed an insertion of three nucleotides (T, C and G) causing the addition of an amino acid valine in the sequence of the LasR protein, in position 53.
ConclusionThe modeling of the resulting LasR protein showed a conformational change in its structure, suggesting that this might be the reason why these isolates are unable to produce biofilm.
Pseudomonas aeruginosa is an opportunistic pathogen frequently associated to nosocomial infections, especially in imunocompromised hosts. This pathogen has also been historically associated to lung infections in patients with cystic fibrosis.1 This microorganism uses several virulence factors to aid in its pathogenicity that will favor infection.2 Among these factors one can highlight the production of biofilm, which is configured as a set of bacterial cells coated by polysaccharide layer, acting as a protection factor of the bacteria against the attack of the antimicrobial and host immune system, favoring the chronicity of the infection.1
The formation of biofilms causes considerable problems in the medical and industrial area, as these structures provoke greater resistance to the treatment with antibiotics and biocides, besides reducing the action of the host immune responses.2 Due to its complexity and the impact of its development, microbial biofilms have been the subject of several studies trying to elucidate how these structures are formed and managed to interact within this community to mitigate the consequences of their formation.3
Biofilm formation is regulated through quorum sensing systems.4 The quorum sensing (QS) is a mechanism of cellular recognition and regulation of expression of many bacterial cell genes, mainly mediated by three interconnected systems, the Las, Rhl and Pqs systems.4 QS is, therefore, important in the formation and maintenance of biofilms by allowing the recognition of the population density, whatever the environmental conditions in which the cells are found, provide the necessary changes to guarantee their survival, through the production of small molecules of communication, the N-acyl-homoserine-lactones (AHL).5
Therefore, the QS plays an important role among the regulation systems of expression of virulence factors that favor bacterial pathogenicity, including biofilm formation.5 In P. aeruginosa, one of the main regulators of QS is the LasR protein, which needs, in order to regulate QS, to bind to its autoinducer, the molecule N-(3-oxododecanoyl)-homoserine lactone (OdDHL).6
The lasR gene codes the transcription factor, which is responsible for the activation of numerous target genes, most of them related to QS in P. aeruginosa. lasR mutations are diverse as well as the phenotypes generated by them.7 Moreover, mutations in lasR gene were recently suggested to be generated during infection, as a result of the strength of lasR transcription.7
In this work, we aimed to investigate biofilm production in multidrug resistant (MDR) and multidrug susceptible (MDS) clinical isolates of P. aeruginosa from different sites of infection and colonization, and describe their clonality. In addition, to sequence PCR products of QS genes for investigating the occurrence of mutations in the LasR protein of non-biofilm producing isolates.
MethodsCharacterization of studyThis was an experimental, laboratory-based study in which 40 clinical isolates of P. aeruginosa stored in the bacterial collection of the Laboratory of Bacteriology and Molecular Biology of the Universidade Federal de Pernambuco (UFPE) were randomly selected, 20 multidrug-susceptible (MDS) and 20 multidrug-resistant (MDR) were analyzed. The study was approved by the Research Ethics Committee of UFPE, registered at CEP/CCS/UFPE under number 009/11. Isolates resistant to at least three different antimicrobial classes, mainly aminoglycosides, penicillins, cephalosporins, carbapenems, and fluoroquinolones were considered multidrug-resistant.8 These isolates were collected from November 2012 to November 2013, from several sites of infection and colonization (Table 1). The identification and analysis of the antimicrobial susceptibility was performed by the automated system (Phoenix – BD®) and sent to the Laboratory of Bacteriology and Molecular Biology of the Universidade Federal de Pernambuco, where they were kept frozen in 20% glycerol at −20°C. The bacteria were reactivated in BHI (Brain Heart Infusion) broth and incubated for 48h at 37°C, and then seeded on cetrimide agar and incubated at 37°C for 24h for further analysis.
Phenotypic characterization of biofilm productionBiofilm production was evaluated according to Stepanovic et al.,9 with modifications. Briefly, 50g/L sucrose was added to BHI broth. The categorization of biofilm production was performed according to the biomass adhered to the bottom of the plate according to the values of the optical densities read at 570nm. The BHI broth was used as negative control (CN) and the P. aeruginosa strain PA01 was used as the positive control (CP), since this strain is recommended as a positive control for biofilm assays.
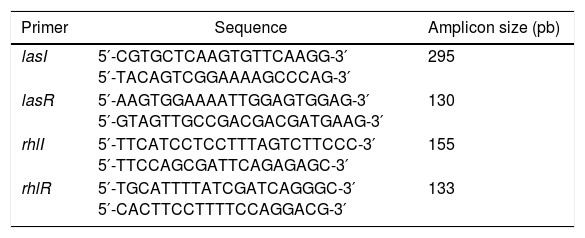
Detection of QS genesDNA extraction from the isolates was performed using the Brazol kit (LGC-Biotechnology) according to the protocol provided by the manufacturer. DNA was quantified by spectrophotometry (Ultraspec 3000; Pharmacia Biotech) over the wavelength range 260–280nm. PCRs were performed to detect the following QS genes: lasR, lasI, rhlI, and rhlR.10,11 The parameters for the amplification were 30 cycles of denaturation at 94°C for 1min, annealing at 52°C for 1min, and extension at 72°C for 1.5min. The PCR products were stained with blue-green (LGC Biotecnologia – São Paulo), submitted to 2% agarose gel electrophoresis and visualized under UV light. The primers are presented in Table 2. The P. aeruginosa strain PA01 was used as the positive control (CP) since this strain is recommended as a positive control for biofilm assays.
Sequences of quorum sensing gene primers.
| Primer | Sequence | Amplicon size (pb) |
|---|---|---|
| lasI | 5′-CGTGCTCAAGTGTTCAAGG-3′ 5′-TACAGTCGGAAAAGCCCAG-3′ | 295 |
| lasR | 5′-AAGTGGAAAATTGGAGTGGAG-3′ 5′-GTAGTTGCCGACGACGATGAAG-3′ | 130 |
| rhlI | 5′-TTCATCCTCCTTTAGTCTTCCC-3′ 5′-TTCCAGCGATTCAGAGAGC-3′ | 155 |
| rhlR | 5′-TGCATTTTATCGATCAGGGC-3′ 5′-CACTTCCTTTTCCAGGACG-3′ | 133 |
Three clinical isolates of P. aeruginosa were randomly selected from the non-biofilm producers for sequencing of the amplicons obtained after QS genes PCR. PCR products were purified with the DNA Wizard® SV gel and PCR clean-up System kit (Promega), according to the manufacturer's protocol and quantified by spectrophotometry. Purified amplicons were submitted to sequencing and the nucleotide sequences were confronted with sequences available in databases using the BlastN and BlastX BlastNetwork algorithms available in the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The nucleotide sequences obtained from each isolate were analyzed and translated into amino acids.12 The multiple sequence alignment was performed in the Clustal Omega program.13,14
Analyses of the amino acid sequences for the three-dimensional modeling of the LasR protein were carried out to assess possible structural differences between these proteins of the non-biofilm-producing isolates and the biofilm-producing strain PAO1. In the PDB database (RCSB Protein Databank) (http://www.rcsb.org/pdb/home/home.do), BlastP was performed between the mature peptide sequence and the structural models deposited in the database, aiming at the selection of the most similar structures to P. aeruginosa LasR protein, which could be used as a model for three-dimensional modeling of the protein. Furthermore, the three-dimensional structures of the candidate proteins were solved through comparative modeling. Multiple alignment between the model structures and quality evaluation were performed using the Protein Structure & Model Assessment tool of the SWISSMODEL Workspace platform (http://swissmodel.expasy.org/).15 Consequently, the quality assessment was based on QMEAN data, which compares the model to deposited structures of the same size and Z-score.16,17 The structures had the various visualization modes generated in the NOC 3.0 program. Using Rampage, were obtained the Ramachandran graph of the structure and its G-factor.18
Molecular typing of the isolatesThe 40 isolates were submitted to molecular typing through technical enterobacterial repetitive intergenic consensus-based PCR (ERIC-PCR) to identify the clonal profile of the strains. The ERIC-PCR reactions were prepared in a total volume of 25μL per tube, comprising: 100ng genomic DNA, 10pmol of the primers (ERIC-1 [5′-ATG TAAGCTCCTGGGGATATACAC-3′]; [5′-AAGTAAGTGACTGGGGTGAGCG-3′]), 1× buffer, 200μM deoxyribonucleotide triphosphate, 1.5mM MgCl2 and 1.0U of Taq DNA polymerase enzyme. The amplification parameters used: initial denaturation at 95°C for 3min, followed by 30 cycles of denaturation at 92°C for 1min, annealing at 36°C for 1min, and extension at 72°C for 8min. After the 30 cycles, a final elongation step of 16min at 72°C was performed. The PCR products were stained with blue-green (LGC Biotecnologia – São Paulo), and submitted to 1.5% agarose gel electrophoresis, visualized under UV light and photodocumented for later analysis of clonal profiles.19
ResultsBiofilm productionIn the quantitative technique for measuring biofilm production, described by Stepanovic et al.,9 a phenotypic test considered a “gold standard” for biofilm detection, 77.5% (31/40) of the isolates were considered biofilm producers, being distributed in the following categories: 42.5% (17/40) weakly adherent, 27.5% (11/40) moderately adherent, and 7.5% (3/40) strongly adherent, as can be observed in Table 3.
Detection of QS genesIn the present study, a high occurrence of QS genes was found, 100% of the isolates presented the lasR, rhlI and rhlR genes and 97.5%, presented the lasI gene, observed in Table 4. In this study, nine clinical isolates of P. aeruginosa (P1A, P9A, P1B, P5B, P7B, P9B, P11B, P19B, and P20B), all presenting different genetic similarity profile, were non-biofilm producers. However, they all presented the genes lasI, lasR, rhlI, and rhlR.
Analysis of the gene and protein sequence of P. aeruginosa PAO1 strain and clinical isolates of non-biofilm producing P. aeruginosa.
| Isolate | Nucleotide sequence | Amino acid sequence |
|---|---|---|
| PAO1 | ATGGCGAGCGACCTTGGATTCTCGAAGATCCTGTTCGGCCTGTTGCCTAAGGACAGCCAGGACTACGAGAACGCCTTCATCGTCGGCAACTAC | MASDLGFSKILFGLLPKDSQDYENAFIVGNY Frame+1 |
| P1A | ATGGCGAGCGACCTTGGATTCTCGAAGATCCTGTTCGGCCTGTTGCCTAAGGACAGCCAGGACTACGAGAACGCCTTCATCGTCG**G*CAACTAC | MASDLGFSKILFGLLPKDSQDYENAFIV#GNY |
| P7B | ATGGCGAGCGACCTTGGATTCTCGAAGATCCTCTTCGGCCTGTTGCCTAAGGACAGCCAGGACTACGAGAACGCCTTCATCGTCGTC**G*CAACTAC | MASDLGFSKILFGLLPKDSQDYENAFIV#GNY |
| P20B | ATGGCGAGCGACCTTGGATTCTCGAAGATCCTGTTCGGCCTGTTGCCTAAGGACAGCCAGGACTACGAGAACGCCTTCATCGTCGTC**G*CAACTAC | MASDLGFSKILFGLLPKDSQDYENAFIV#GNY |
* Regions where nucleotide insertions (T, C and G) occurred, respectively, in clinical isolates of non-biofilm producing P. aeruginosa.
# Regions where amino acid valine insertion occurred, in clinical isolates of non-biofilm producing P. aeruginosa.
When comparing the lasR gene sequences of three non-biofilm producing isolates (P1A, P7B and P20B), it was evidenced, through the alignment of sequences with the biofilm-producing strain P. aeruginosa PAO1 (default biofilm producing strain), the insertion of three nucleotides (T, C and G) between the positions 1558327–1558330 in the lasR gene sequences (Table 4). When aligning these three sequences with the strain of P. aeruginosa PAO1 by NCBI BlastP tool, it was observed that the insertions led to the addition of the amino acid valine at LasR protein sequence, at position 53, frame 2.
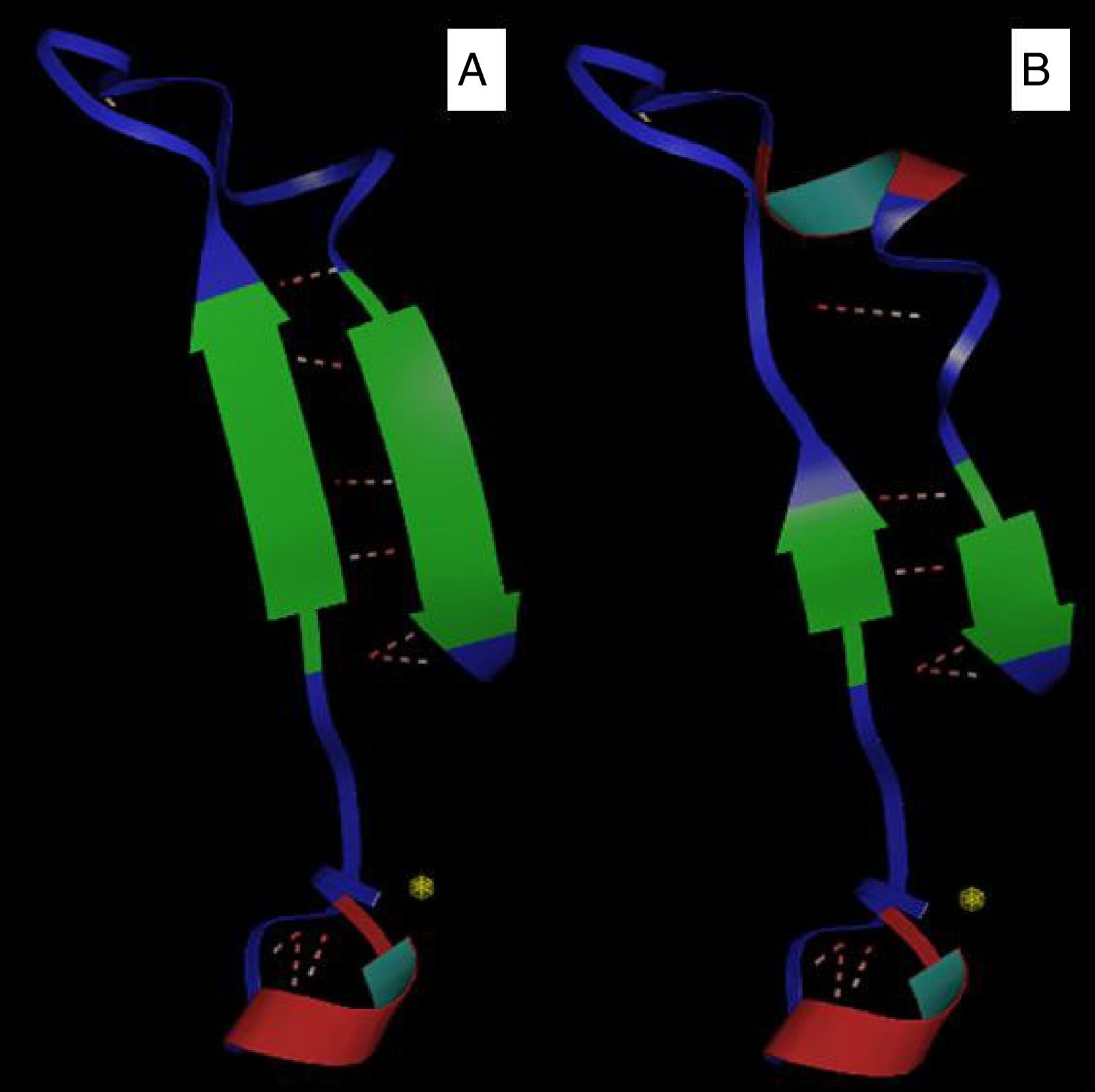
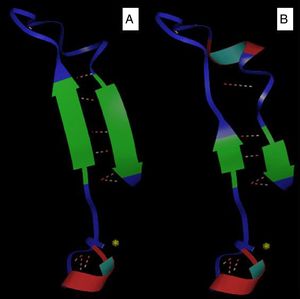
Bioinformatics analysisThe comparative modeling of the LasR variant was based on the use of a known model (with experimentally determined structure), aligned with the sequences for which the tertiary structure was desired. Furthermore, this analysis allowed a visualization of the general architecture of these carbohydrate binding proteins. In this study, the models were constructed from the model template 2uv0 and 3jpu, both of P. aeruginosa, with 1.8Å and 2.3Å resolution, respectively, available in the Protein Data Bank (PDB), which shows the best similarity with the proteins of the analyzed species. Thus, these sequences were submitted to homology-based computational modeling, presenting 100% identity values after alignment against the model and strains P. aeruginosa PAO1 and P1A isolate (non-biofilm producer). Three-dimensional modeling demonstrated that the insertion of the amino acid valine into the protein sequence of the P1A isolate generated a change in the protein structure, reducing the antiparallel beta sheets and altering the structure of the alpha cleavage (Fig. 1A and B).
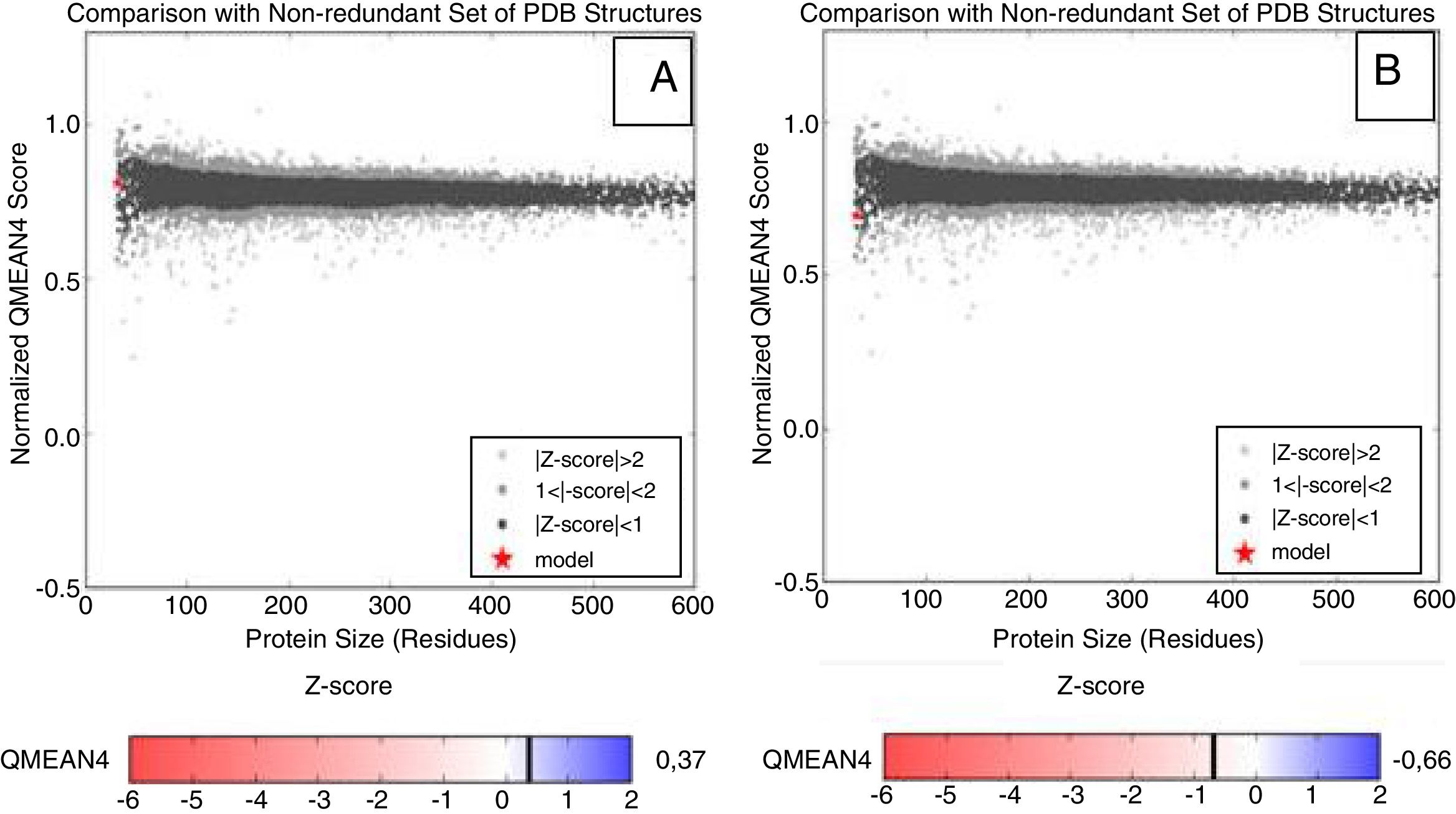
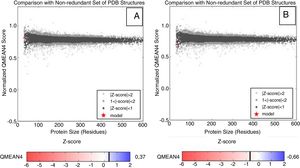
The reliability analysis of the proposed models presented values within the reliability standards, with QMEAN scores varying within the spectrum −0.66 to 0.37 (values closer to one represent models with higher quality). Although negative Z-scores are an indication of a good model, the plot of the model should also be considered in the graph of the scores of all protein structures deposited in the PDB, which must be located inside the gray (light or dark) and black areas. Based on this analysis, the different proteins modeled are within the area referred to the desired quality standards, and the LasR protein was modeled for the clinical isolate of P. aeruginosa P1A, which is non-biofilm producer, plotted more externally to the graph (Fig. 2A and B).
Representative Z-score graphs of the generated models, compared to X-ray (gray) and magnetic (dark gray) crystallography models deposited in the PDB. The dots in red refer to the generated models for the P. aeruginosa LasR protein. (A) P. aeruginosa PAO1 (standard biofilm producing-strain) and (B) P. aeruginosa P1A (clinical isolate non-biofilm producer), obtained through comparative modeling.
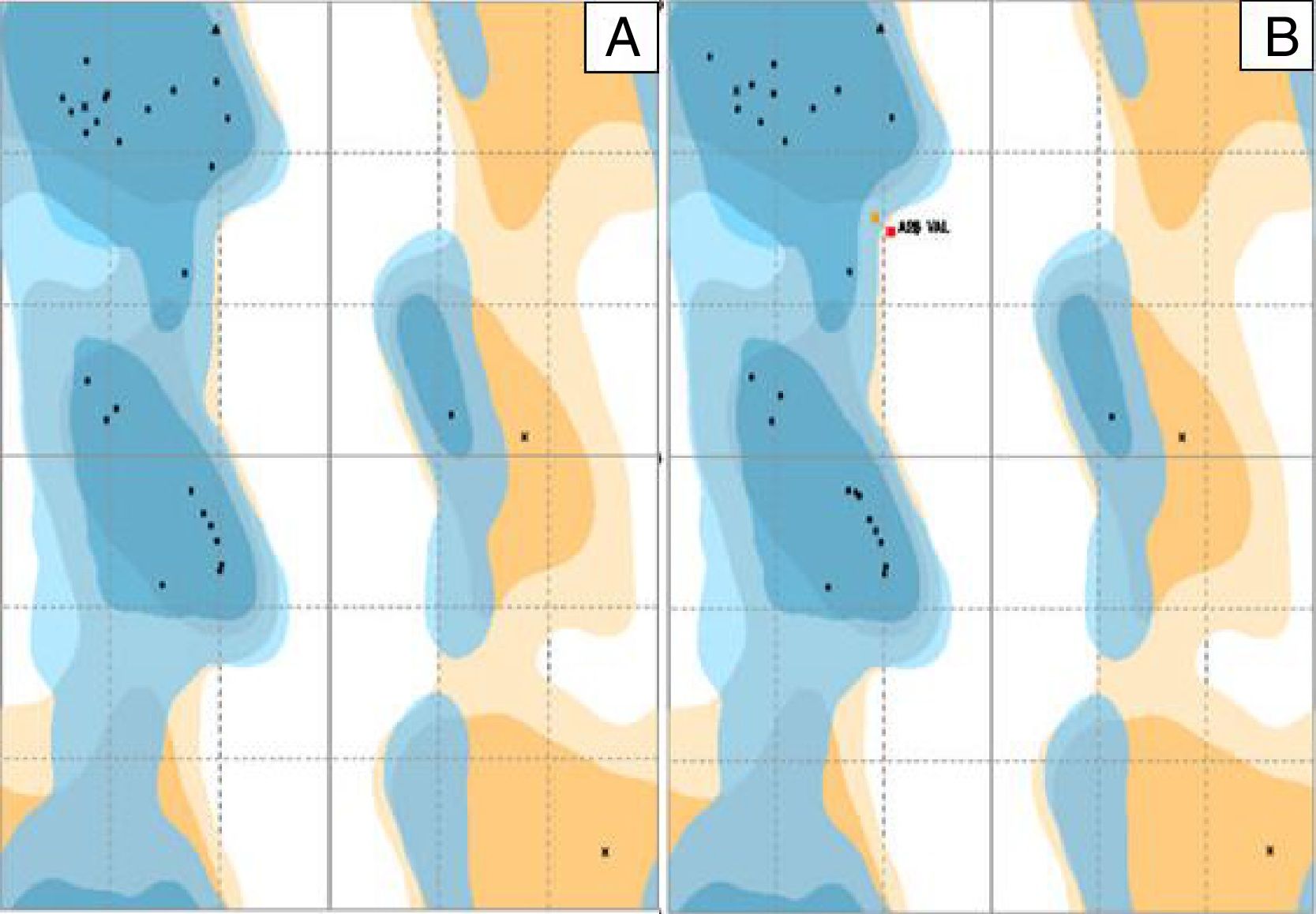
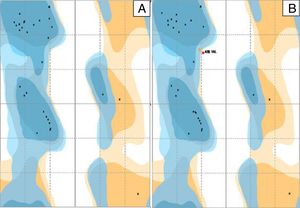
The distribution of the amino acid residues in the Ramachandran graph, which evaluates the stereochemical quality of the structure, revealed that 98% and 93.3% of the residues were in the most favorable energy regions for a good quality model for P. aeruginosa PAO1 (standard biofilm producing strain) and P. aeruginosa P1A (clinical non-biofilm producing isolate), respectively (Fig. 3A and B). In the graph of P. aeruginosa P1A, there are amino acid residues in the energetically unfavorable regions, due to the presence of the amino acid valine, differing from the results described by Chowdhury et al.,20 in which no amino acid residue was in the most unfavorable energy regions of the Ramachandran chart.
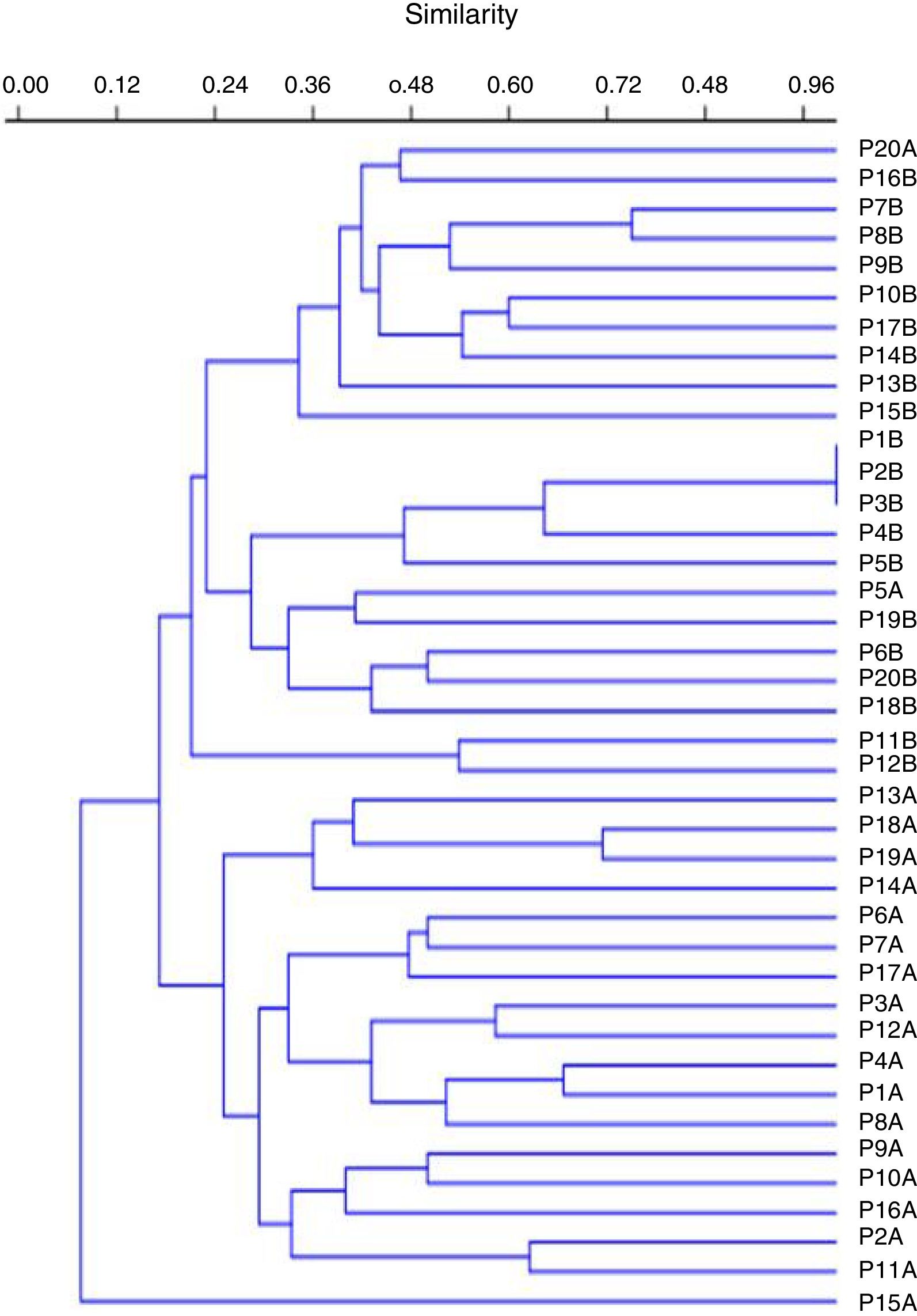
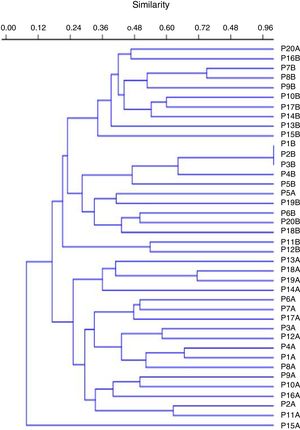
Molecular typingThe genotyping of clinical isolates of P. aeruginosa by ERIC-PCR showed 38 distinct genetic profiles, with similarity ranging from 24% to 100% (Fig. 4).
DiscussionThe biofilm quantification test proved to be effective in the detection of biofilm production by the clinical isolates analyzed, since it was able to detect biofilm production by P. aeruginosa strain PAO1, used as positive control in the test. Similar data were observed by Lima et al.,21 showing that 75% of the clinical isolates of P. aeruginosa were biofilm producers. Additionally, in a study conducted by Perez et al.,22 in which biofilm production was observed in 68% (50/74) of the clinical isolates of P. aeruginosa, being distributed in the following categories: 96% (48/50) weakly adherent and 4% (2/50) moderately adherent. In the study of Perez et al.,23 biofilm production was present in 93.4% (85/91), being distributed in the following categories: 60% (51/85) poorly adherent, 25.9% (22/85) moderately adherent, and 14.1% (12/85) strongly adherent.
Furthermore, among the isolates classified as biofilm producers in the present study, 48.4% (15/31) were MDR and 51.6% (16/31) were MDS, suggesting that, for these isolates, biofilm production was not relevant for antimicrobial resistance, differing from the results found by Perez et al.,22 where all isolates of P. aeruginosa producing metallo-β-lactamases (MβL) produced biofilm.
As for the occurrence of the genes of QS, our data are similar to those found by Perez et al.,23 where 90.1% (82/91) of the analyzed isolates presented all the genes. By contrast, the results reported by Karatuna et al.24 showed that QS genes detection were not as high, as those reported in our study detection (81.25% for the lasI and lasR genes, 68.75% for the rhlI gene, and 62.5% for the rhlR gene).
In the present study, the P. aeruginosa isolate unassembled only for the lasI gene was a strong biofilm producer. In the same way, in the study conducted by Perez et al.,23 an unassembled isolate for the lasR gene, was biofilm producer. These facts can be explained by the in vivo compensation for the presence of the rhlI/rhlR QS system, as observed in P. aeruginosa lasR-mutant which showed no reduction in its virulence during a corneal infection process.10
In a study conducted by Sabharwal et al.,25 in India, the occurrence of lasI, lasR, rhlI, and rhlR genes of P. aeruginosa was 75% for the lasI and lasR genes, 41.6% for the rhlI gene, and 58.3% for the rhlR gene. Accordingly, similar data were found in the study by Aboushleib et al.,26 in Egypt, where the QS genes occurrences were 48% for the lasI gene, 40% for the lasR and rhlI genes, and 36% for the rhlR gene. Analysis of these studies revealed that genotypically positive and phenotypically negative isolate sequences appear to carry several mutations in the QS genes.
To the best of our knowledge, despite the report of numerous lasR mutations, the mutation reported herein had never been described elsewhere. In the study carried out by Senturk et al.,27 four isolates of P. aeruginosa were non-biofilm producers, despite the presence of these QS genes. The authors attributed the lack biofilm production to the mutations detected by sequencing, although they did not report the types of mutations found. Bjarnsholt et al.28 evaluated the occurrence of mutations in the QS genes of 238 P. aeruginosa isolates obtained from cystic fibrosis patients. Mutations in the lasR and rhlR genes were found and these isolates showed decreased virulence, similar to those found in the present study in which isolates that had mutations in the lasR gene were non-biofilm producers.
By contrast, Aboushleib et al.26 found mutations in the lasI, lasR, rhlI, and rhlR genes in both biofilm producer and non-producer isolates.
Moreover, in the study by Feltner et al.,29 the lasR gene of 2583 cystic fibrosis isolates of P. aeruginosa was sequenced, and 580 sequences encoded proteins that differed from conserved LasR proteins. The authors showed that about one-sixth of 31 LasR variant proteins were functional, including three with nonsense mutations. In addition, in lasR-null isolates, LasR-dependent were, nevertheless, expressed. The remaining 23 isolates showed non-functional LasR variants. Although the findings did not describe the mutation found in our study, it is possible that this mutation did also generate a non-functional LasR variant, which would explain the lack of biofilm production.
In addition, the lasR gene is located at the position 1558171–1558890 of the genome of P. aeruginosa and is franchised by genes PA1429 and rsal,7 responsible for cytoplasmic LasR protein coding. This protein belongs to the family of transcriptional regulators LuxR-type and is related to the regulation of transcriptional regulatory region, autoinducer region (area where the connection with the autoinducers occurs) and the region of the DNA-binding residues.30 The mutation described in our study was in the first two regions, which could therefore prevent biofilm formation.
Bottomley et al.30 observed through modeling that the region of insertion of the amino acid valine corresponds to the region of the beta 2 leaf. In addition, the analysis of the modeling reveals that the atomic interactions between the protein LasR and its autoinducer N-(3-oxododecanoyl)-homoserine lactone (OdDHL) occur in the N-terminal domain of the LasR protein, through hydrogen bonds. The amino acid residues found to be involved in the interactions with the autoinducer were tyrosine-56, arginine-61, aspartate-73, threonine-115 and serine-129. These residues bind concurrently and cause protein folding, leading to dimerization of LasR, thus allowing DNA binding to the promoter and consequent transcriptional activation of QS controlled genes. These amino acids, detected by computational analysis, provide information on the binding affinities of the amino acid residues to the specific ligands.30 Based on this analysis, it can be suggested that the valine insertion in position 53 of the LasR protein prevented the binding of the amino acid tyrosine located in position 56 to form the hydrogen bond with the autoinducer, preventing activation of the protein biofilm formation.
Our results corroborate the findings of Bjarnsholt et al.,28 in which amino acids tyrosine-56 and threonine-75 strongly interact with the autoinducer, and the occurrence of mutations in this region of the lasR protein would impair autoinducer binding.
A great genetic diversity was observed in the clinical isolates of P. aeruginosa analyzed in this study, as found in studies involving patients with severe or chronic infections, who present favorable conditions for the occurrence of non-lethal mutations.31,32 Similar data were also observed by Cavalcanti et al.,33 who reported replacement of the SPM-1 P. aeruginosa clone by a variety of genetically distinct P. aeruginosa isolates in a university hospital in Recife, Brazil. Besides that, the presence of clones with phenotypic profiles of distinct biofilm production evidences the existence of regulation mechanisms of the expression of QS genes related to biofilm production. Of concern is the high biofilm production by these isolates, even by multridrug sensitive isolates, which may make it difficult to treat the infections caused by them. The three-dimensional models were satisfactory in the evaluation of the regulation of the proteins related to biofilm formation, and may, therefore, be used in other analyses. However, further studies including a larger number of clinical isolates of P. aeruginosa will be necessary to elucidate the relationship between this mutation found and non-biofilm producing clinical isolates of P. aeruginosa.
FundingThis research was in part supported by National Council for Scientific and Technological Development (CNPq) and the Coordination for the Improvement of Higher Education Personnel (CAPES).
Conflicts of interestThe authors declare no conflicts of interest.