Oligoadenylate synthetases play an important role in the immune response against dengue virus. Single nucleotide polymorphisms in the oligoadenylate synthetases genes are known to affect oligoadenylate synthetases activity and are associated with outcome of viral infections. Polymorphisms in the OAS1 SNPs (rs1131454), OAS2 SNPs (rs1293762, rs15895 and rs1732778) and OAS3 SNPs (rs2285932 and rs2072136) genes were studied using PCR followed by restriction fragment length polymorphism methods in 30 patients for dengue infection and 40 control group who have no documented evidence of symptomatic dengue. An increase in the frequency of OAS2 gene rs1293762 SNP G/T heterozygotes (p=0.012), decrease in the frequency of SNP G/G homozygotes (p=0.005) and decrease in the frequency of OAS2 gene rs1732778 SNP G/G homozygotes (p=0.000017) and A/A homozygotes (p=0.0000012) were observed among the dengue patients compared with control group. Our results suggest that OAS2 haplotypes are associated with differential susceptibility to clinical outcomes of dengue virus infection.
Dengue has emerged as a global health problem, as evidenced by a series of epidemics throughout the tropical, subtropical and temperate regions of the world.1 Dengue virus (DENV), member of the flavivirus genus of the Flaviviridae family, is an enveloped and mosquito borne virus that contains a single-stranded, positive sense RNA genome. DENV infection can lead to dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS). The WHO has reported that DENV infection causes an estimated 50–100 million cases of DF and several hundred thousand cases of DHF/DSS annually around the world.2 The pathophysiology of dengue virus (DENV) infection is multifactorial involving complex interactions among viral and host factors such as human leukocyte antigens and cytokines.3
The 2′-5′-oligoadenylate synthetase (2′-5′-OAS) endoribonuclease L (RNase L) pathway is an important component of the antiviral intracellular innate immune response in mammalian cells.4 OAS gene expression is induced by type I interferons and type II interferons. Viral double-stranded RNA or single-stranded RNA can activate 2-5-OAS to polymerize short 2′-5′-linked oligoadenylate. 2′-5′-Linked oligoadenylate binds to latent RNase L, which causes its dimerization and activation. The activated RNase L cleaves both cellular RNA and viral RNA molecules, resulting in degradation of protein synthesis and inhibition of virus replication.5 In the present study, the possible association of six SNPs distributed across the three human OAS genes with predisposition to severe dengue virus infection-induced disease was investigated.
Blood samples were obtained from dengue patients, who gave informed consent for participation in the research, which was approved by the Ethical Committee of the Institute of Regional Medical Research Centre, Port Blair. A total of 30 samples were obtained from patients with dengue virus infection and compared with random control samples from 40 individuals living in the Andaman Islands with no information about dengue virus infection.
DNA was extracted by using the QIAmp DNA Blood Maxi Kit (Qiagen) and stored at −20°C before use. Polymerase chain reaction and restriction fragment length polymorphism analysis was performed to genotype OAS1 SNP rs1131454, OAS2 SNPs rs1293762, rs15895, and rs1732778 and the OAS3 SNPs rs2285932 and rs2072136 in dengue samples and control samples as previously described.5 The restriction products were visualized on either 2% agarose gels. Individual SNP allele and genotype frequencies and haplotype frequencies were compared between patients with dengue infection and control groups using Chi-square and Fisher exact test using (EpiInfo 7 software – www.cdc.gov/epiinfo/). Differences between groups were considered significant at p<0.05. Permutation tests using the EM algorithm were applied to analyze pair-wise linkage disequilibrium between SNPs.
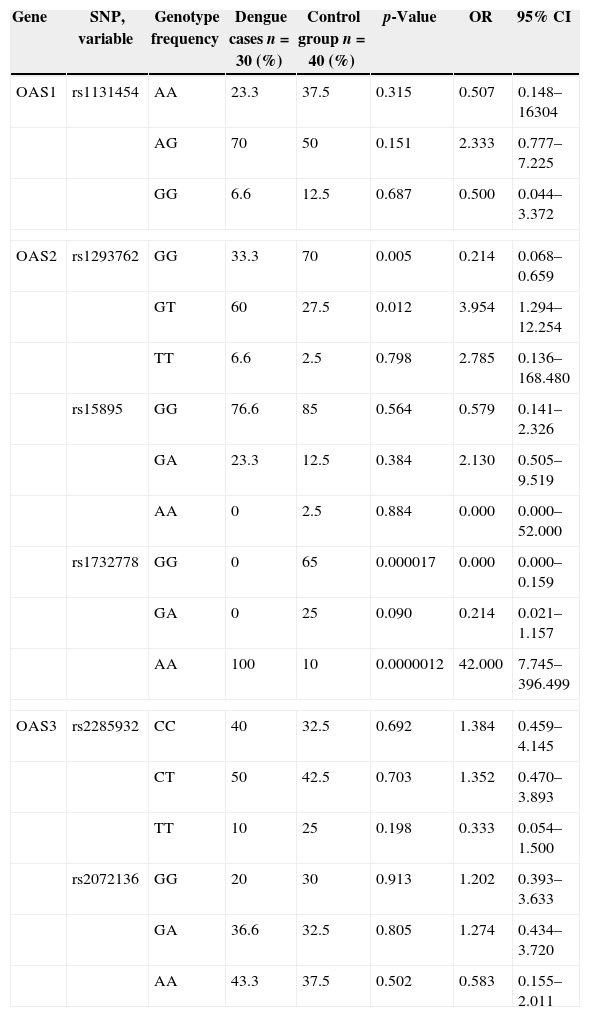
A total of six SNPs located in different regions of the OAS1 (1SNPs), OAS2 (3SNPs) and OAS3 (SNPs2) genes were selected. OAS SNPs were genotyped in 30 DNA samples from dengue patients and 40 DNA samples from control group. Statistically significant differences in the genotype between the dengue cases and healthy controls were observed. Characteristics of these SNPs in multiple OAS1, OAS2, and OAS3 gene transcripts are indicated in Table 1.
Statistical association of single nucleotide polymorphisms for dengue cases compared to healthy individuals.
| Gene | SNP, variable | Genotype frequency | Dengue cases n=30 (%) | Control group n=40 (%) | p-Value | OR | 95% CI |
|---|---|---|---|---|---|---|---|
| OAS1 | rs1131454 | AA | 23.3 | 37.5 | 0.315 | 0.507 | 0.148–16304 |
| AG | 70 | 50 | 0.151 | 2.333 | 0.777–7.225 | ||
| GG | 6.6 | 12.5 | 0.687 | 0.500 | 0.044–3.372 | ||
| OAS2 | rs1293762 | GG | 33.3 | 70 | 0.005 | 0.214 | 0.068–0.659 |
| GT | 60 | 27.5 | 0.012 | 3.954 | 1.294–12.254 | ||
| TT | 6.6 | 2.5 | 0.798 | 2.785 | 0.136–168.480 | ||
| rs15895 | GG | 76.6 | 85 | 0.564 | 0.579 | 0.141–2.326 | |
| GA | 23.3 | 12.5 | 0.384 | 2.130 | 0.505–9.519 | ||
| AA | 0 | 2.5 | 0.884 | 0.000 | 0.000–52.000 | ||
| rs1732778 | GG | 0 | 65 | 0.000017 | 0.000 | 0.000–0.159 | |
| GA | 0 | 25 | 0.090 | 0.214 | 0.021–1.157 | ||
| AA | 100 | 10 | 0.0000012 | 42.000 | 7.745–396.499 | ||
| OAS3 | rs2285932 | CC | 40 | 32.5 | 0.692 | 1.384 | 0.459–4.145 |
| CT | 50 | 42.5 | 0.703 | 1.352 | 0.470–3.893 | ||
| TT | 10 | 25 | 0.198 | 0.333 | 0.054–1.500 | ||
| rs2072136 | GG | 20 | 30 | 0.913 | 1.202 | 0.393–3.633 | |
| GA | 36.6 | 32.5 | 0.805 | 1.274 | 0.434–3.720 | ||
| AA | 43.3 | 37.5 | 0.502 | 0.583 | 0.155–2.011 | ||
An increased frequency of OAS2 gene rs1293762 SNP G/T heterozygotes was observed among dengue patients when compared with control group. The difference was statistically significant (p=0.012, OR=3.954, 95% CI: 1.294–12.254). The frequency of OAS2 gene r1293762 SNP G/G homozygotes was significantly decreased among dengue patients compared to control group (p=0.005, OR=0.124, 95% CI: 0.068–0.659). Likewise, the frequency of OAS2 gene rs1732778 SNP G/G homozygotes and A/A homozygotes was significantly decreased among dengue patients compared to control group (p=0.000017, OR=0.000, 95% CI: 0.000–0.159 and p=0.0000012, OR=42.000, 95% CI: 7.745–396.49, respectively). No significant association with dengue infection was observed for OAS1 gene rs1131454 SNP homozygotes AA (p=0.315), GG (p=0.687) and heterozygote AG (p=0.151); OAS3 gene rs2285932 SNP homozygotes CC (p=0.692), TT (p=0.198) and CT (p=0.703) and rs2072136 SNP homozygotes GG (p=0.913), AA (p=0.502) and GA (p=0.805) SNPs.
The severity of DENV infection is determined by complex factors including magnitude of DENV replication, expression of cytokines, activation and proliferation of immune cells, host genetic factors, etc.6,7 As patients with DHF/DSS tend to have higher viremia titers than patients with dengue fever, DENV levels appear to be associated with the severity of dengue diseases.8,9 Oligoadenylate synthetase is well known as an IFN induced antiviral pathway, which plays a critical role in innate immunity, controlling the outcome of virus production. The OAS3 protein preferentially catalyzes the synthesis of dimeric 2′-5′-linked Oligoadenylate that binds to RNase L with lower affinity and induces less activation than the trimeric and longer oligomers synthesized by the OAS1 and OAS2 proteins.10 OAS3 has been reported to have antiviral activity against the alphavirus chikungunya.11
The anti-DEN activities of these three OAS isoforms correlated with their ability to trigger RNase L activation in DENV infected cells. However, OAS1 p42/p46 and OAS3 p100 are likely to contribute to host defense against DENV infection and play a role in determining the outcomes of DEN disease.12
2′-5′-OAS controls DENV replication and provides insights into the antiviral capacity of various members of the OAS family against DENV replication. The antiviral effects of human OAS1 p42, OAS1 p46, and OAS3 p100 correlate with their abilities to trigger RNase L activation in DENV-2 infected cells.12 It would be interesting to determine whether these three OAS isoforms also contribute to host defense against other flaviviruses and whether they are candidate human genetic factors for determining human susceptibility and severity of DENV-related diseases.
Associations between two SNPs located within the OAS2 gene and the outcome of dengue virus infection were detected in the present study. However, in the present cohort there was no evidence of a significant association between DENV infection and OAS1 SNP rs1131454, OAS3 SNPs rs2285932 and rs2072136. Of these OAS genes SNPs were deviated from Hardy–Weinberg equilibrium (HWE), but were not significantly associated with dengue virus infection. Another study13 suggested that Hardy–Weinberg equilibrium was not significantly associated with West Nile virus infection or disease progression from 360 SNPs distributed across 86 genes.
An association was noticed between the OAS1 gene rs2660 SNP located in the 3-untranslated region of p46 and the outcome of infection with another member of the Flaviviridae family namely hepatitis C virus.14 In the present study no association was found between any of the OAS1 gene SNP and OAS3 SNPs tested and human susceptibility to dengue virus. A recent study showed that OAS1-OAS3-OAS2 haplotypes were associated with differential susceptibility to clinical outcomes of dengue infection.15 In the present study data obtained from the analysis of OAS gene SNPs suggest that variation in this component of the innate immune response could contribute to the outcome of dengue virus infection. Nonetheless, data from additional cohorts are needed to further validate these results; identification of genetic variants that control the innate immune response will provide an increased understanding the mechanisms of viral pathogenesis.
Conflicts of interestThe authors declare no conflicts of interest.
The authors are thankful to the Indian Council of Medical Research for providing financial grants for the study.