A better understanding of the antimicrobial susceptibility, carriage of virulence determinants and molecular characteristics of Staphylococcus aureus isolates associated with skin and soft tissue infections (SSTIs) may provide further insights related to clinical outcomes with these infections. From January 2012 to September 2013, a total of 128 non-duplicate S. aureus isolates were recovered from patients with SSTIs. All 128 S. aureus SSTI isolates carried at least five virulence genes tested. Virulence genes detected among at least 70% of all tested isolates included hld (100%), hla (95.3%), icaA (96.9%), clf (99.2%), sdrC (79.7%), sdrD (70.3%), and sdrE (72.7%). The prevalence of MRSA isolates with 10 virulence genes tested (54.4%, 31/56) was significantly higher than that among MSSA isolates (35.2%, 25/71) (p<0.05). The positive rates of seb, sen, sem, sdrE and pvl among MRSA isolates were significantly higher than among MSSA isolates (p<0.05). ST7 and ST630 accounting for 10.9% were found to be the predominant STs. The most prevalent spa type was t091 (8.6%). MRSA-ST59-SCCmec IV was the most common clone (12.3%) among MRSA isolates whereas among MSSA isolates the dominant clone was MSSA-ST7 (15.5%). Six main clonal complexes (CCs) were found, including CC5 (52.3%), CC7 (11.7%), CC59 (8.6%), CC88 (6.3%), CC398 (4.7%), and CC121 (3.1%). A higher carriage of seb and sec was found among CC59 isolates. In comparison to CC5 and CC7 isolates, those with the highest carriage rates (>80.0%) of sdrC and sdrD, CC59 isolates had lower prevalence of these two virulence genes. All CC59 isolates were susceptible to gentamicin and trimethoprim/sulfamethoxazole, while CC5 and CC7 isolates had resistance rates to these two antimicrobials of 25.4% and 20.9%, and 40.0% and 40.0%, respectively. The resistance rates for tetracycline, clindamycin, and erythromycin among CC5 isolates were lower than among CC7 and CC59 isolates. In conclusion, the molecular typing of S. aureus SSTI isolates in the present study showed considerable heterogeneity. ST7 and ST630 became prevailing clones. Different S. aureus clones causing SSTIs were associated with specific antimicrobial resistance and virulence gene profiles.
Staphylococcus aureus, particularly methicillin-resistant S. aureus (MRSA), is an important human pathogen responsible for many infectious diseases including skin and soft tissue infections (SSTIs), foreign-body infections, pneumonia, septic arthritis, endocarditis, osteomyelitis, sepsis, and bloodstream infections in both hospital and community settings.1 The ability of this clinically important pathogen to successfully persist within the hosts is largely due to the carriage of a battery of virulence factors which promote adhesion, acquisition of nutrients, and evasion of host immunologic responses.2,3 Some S. aureus isolates also produce one or more additional exoproteins, such as toxic shock syndrome toxin-1 (TSST-1), staphylococcal enterotoxins (SEs), exfoliative toxins (ETs), and leukocidins.2–4 Recently, Panton-Valentine leukocidin (pvl) encoded by two contiguous and cotranscribed genes (lukS-PV and lukF-PV) is an important virulence factor for community-acquired MRSA (CA-MRSA) affecting individuals without apparent risk factors for hospital acquisition.5,6S. aureus is the most common bacterial pathogen identified from SSTIs.7 SSTIs caused by MRSA is associated with a high incidence of treatment failure and recurrence.8 A better understanding of the antimicrobial susceptibility, carriage of virulence determinants, and molecular characteristics of S. aureus isolates associated with SSTIs may provide further insights related to clinical outcomes of these infections. Molecular typing has proved to be an important tool to investigate MRSA epidemiology. Pulsed-field gel electrophoresis (PFGE) patterns, SCCmec typing, spa typing, and multi-locus sequence typing (MLST) have been proven useful for monitoring the evolutionary process of pandemic MRSA clones.1 In China, ST239-MRSA-III is a predominant MRSA clone among adults, while ST59-MRSA-IV is the most prevalent clone among children.9,10 In a previous study we investigated the molecular typing of S. aureus isolated from patients with SSTIs at our hospital from December 2002 to June 2008 and found that ST239, ST1018, ST59, ST7, and ST88 were the most prevalent sequence types.11 A shift of important clones has been observed in several studies.12–14 A report from China found a rapid change of MRSA over a 15-year period at a tertiary care hospital, when the ST239-MRSA-III-t037 clone was replaced by the emerging ST239-MRSA-III-t030 clone.15 Understanding the shift of important clones at the local and international levels is of great significance. To understand the shift of S. aureus clones associated with SSTIs, the present study aimed to investigate the antimicrobial susceptibility, carriage of virulence determinants, and molecular characteristics of S. aureus isolates associated with SSTIs at the hospital in 2012–2013.
Materials and methodsCollection of clinical isolates and S. aureus confirmationFrom January 2012 to September 2013, a total of 128 non-duplicate S. aureus isolates (single isolate per patient) were collected at The First Affiliated Hospital of Wenzhou Medical University, China from pus samples of hospitalized patients with SSTIs. Lesions requiring incision and drainage or with spontaneously draining purulent fluid, carbuncles, furuncles, boils, cellulitis with purulent drainage, chronic ulcer, and deep wounds were included. S. aureus isolates from patients with SSTIs with clinical signs and symptoms of infection such as increased white blood cell counts, fever, local redness, swelling, and exudate were considered invasive isolates and included for investigation. Isolates were identified as S. aureus using Gram stain, positive catalase and coagulase test results, and Vitek microbiology analyzer (bioMérieu, Marcy l’Etoile, France). S. aureus ATCC25923 was used as a control strain.
Ethics statementThis study was approved by the Institutional Ethics Review Board of The First Affiliated Hospital of Wenzhou Medical University. All patients provided written informed consent for this study. The written informed consents were also obtained from the next of kin, caretakers, or guardians on behalf of the minors/children enrolled.
Antimicrobial susceptibility testingS. aureus susceptibility to penicillin (10 units), erythromycin (15μg), clindamycin (2μg), rifampicin (5μg), tetracycline(30μg), linezolid (30μg), mupirocin (5μg), quinupristin/dalfopristin (15μg), trimethoprim/sulfamethoxazole (1.25/23.75μg), gentamicin (10μg), ciprofloxacin (5μg), Chloramphenicol (30μg), and nitrofurantoin (300μg) were determined using disc diffusion test recommended by the Clinical and Laboratory Standards Institute (CLSI).16 All discs were obtained from Oxoid Ltd. Vancomycin MICs for S. aureus isolates were determined by agar dilution method. Interpretive standards for the antimicrobial susceptibility test and D-test for tested S. aureus isolates were in accordance with the guidelines provided by CLSI.16 Susceptibility of S. aureus to mupirocin was determined by disc diffusion, with a zone diameter ≥14mm on a 5μg disc indicating susceptibility as described previously.17,18S. aureus ATCC 25923 and Escherichia coli ATCC25922 were used as reference strains for antimicrobial susceptibility testing.
DNA extractionS. aureus isolates tested were cultured on blood agar overnight at 35°C. Then, three to four bacterial colonies were suspended and incubated in 150μL sterile distilled water with lysostaphin (1mg/mL) (Sangon, China) at 37°C for 1h. Finally, DNA was extracted following the instructions of the Genomic DNA Extraction kit (Sangon, China). The extracted DNA was stored at −20°C and prepared for PCR detection.
Identification of MRSA isolates and pvl detectionA multiplex PCR protocol was used for simultaneous amplification of mecA, 16S rRNA, and pvl genes as described previously.19 MRSA isolates harbouring mecA were confirmed using MRSA N315 and pvl-positive MRSA isolate identified in our previous study as positive control strains.20
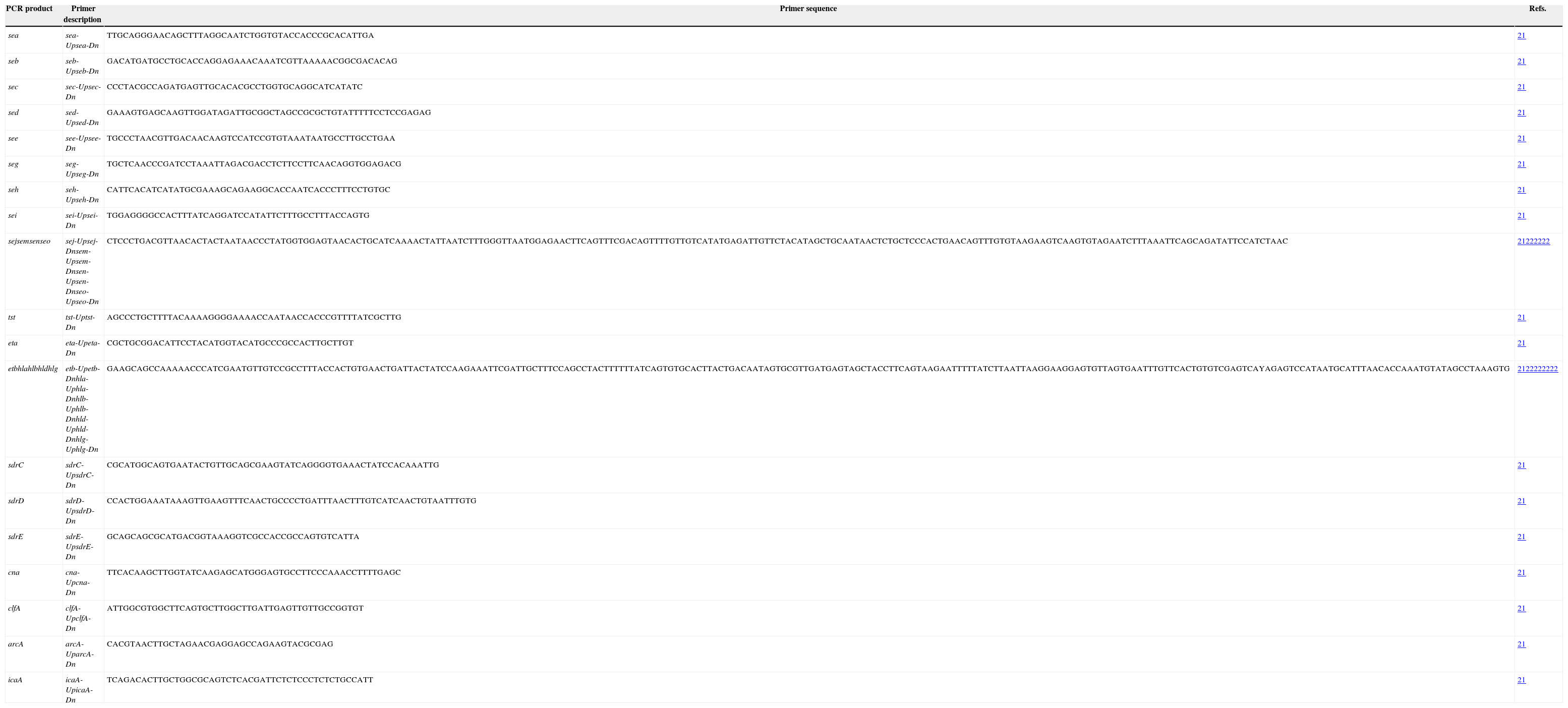
Detection of virulence genesVirulence genes, including toxins (sea, seb, sec, sed, seg, seh, sei, sej, seo, sen, sem, edin, hla, hlb, hld, hlg, tst, eta, etb), adhesins (clfA, cna, sdrC, sdrD, and sdrE), icaA and arcA were identified using PCR assays with primers and conditions previously described.21,22 PCR primers used for PCR assays were shown in Table 1.
PCR primers used for PCR assays.
| PCR product | Primer description | Primer sequence | Refs. |
|---|---|---|---|
| sea | sea-Upsea-Dn | TTGCAGGGAACAGCTTTAGGCAATCTGGTGTACCACCCGCACATTGA | 21 |
| seb | seb-Upseb-Dn | GACATGATGCCTGCACCAGGAGAAACAAATCGTTAAAAACGGCGACACAG | 21 |
| sec | sec-Upsec-Dn | CCCTACGCCAGATGAGTTGCACACGCCTGGTGCAGGCATCATATC | 21 |
| sed | sed-Upsed-Dn | GAAAGTGAGCAAGTTGGATAGATTGCGGCTAGCCGCGCTGTATTTTTCCTCCGAGAG | 21 |
| see | see-Upsee-Dn | TGCCCTAACGTTGACAACAAGTCCATCCGTGTAAATAATGCCTTGCCTGAA | 21 |
| seg | seg-Upseg-Dn | TGCTCAACCCGATCCTAAATTAGACGACCTCTTCCTTCAACAGGTGGAGACG | 21 |
| seh | seh-Upseh-Dn | CATTCACATCATATGCGAAAGCAGAAGGCACCAATCACCCTTTCCTGTGC | 21 |
| sei | sei-Upsei-Dn | TGGAGGGGCCACTTTATCAGGATCCATATTCTTTGCCTTTACCAGTG | 21 |
| sejsemsenseo | sej-Upsej-Dnsem-Upsem-Dnsen-Upsen-Dnseo-Upseo-Dn | CTCCCTGACGTTAACACTACTAATAACCCTATGGTGGAGTAACACTGCATCAAAACTATTAATCTTTGGGTTAATGGAGAACTTCAGTTTCGACAGTTTTGTTGTCATATGAGATTGTTCTACATAGCTGCAATAACTCTGCTCCCACTGAACAGTTTGTGTAAGAAGTCAAGTGTAGAATCTTTAAATTCAGCAGATATTCCATCTAAC | 21222222 |
| tst | tst-Uptst-Dn | AGCCCTGCTTTTACAAAAGGGGAAAACCAATAACCACCCGTTTTATCGCTTG | 21 |
| eta | eta-Upeta-Dn | CGCTGCGGACATTCCTACATGGTACATGCCCGCCACTTGCTTGT | 21 |
| etbhlahlbhldhlg | etb-Upetb-Dnhla-Uphla-Dnhlb-Uphlb-Dnhld-Uphld-Dnhlg-Uphlg-Dn | GAAGCAGCCAAAAACCCATCGAATGTTGTCCGCCTTTACCACTGTGAACTGATTACTATCCAAGAAATTCGATTGCTTTCCAGCCTACTTTTTTATCAGTGTGCACTTACTGACAATAGTGCGTTGATGAGTAGCTACCTTCAGTAAGAATTTTTATCTTAATTAAGGAAGGAGTGTTAGTGAATTTGTTCACTGTGTCGAGTCAYAGAGTCCATAATGCATTTAACACCAAATGTATAGCCTAAAGTG | 2122222222 |
| sdrC | sdrC-UpsdrC-Dn | CGCATGGCAGTGAATACTGTTGCAGCGAAGTATCAGGGGTGAAACTATCCACAAATTG | 21 |
| sdrD | sdrD-UpsdrD-Dn | CCACTGGAAATAAAGTTGAAGTTTCAACTGCCCCTGATTTAACTTTGTCATCAACTGTAATTTGTG | 21 |
| sdrE | sdrE-UpsdrE-Dn | GCAGCAGCGCATGACGGTAAAGGTCGCCACCGCCAGTGTCATTA | 21 |
| cna | cna-Upcna-Dn | TTCACAAGCTTGGTATCAAGAGCATGGGAGTGCCTTCCCAAACCTTTTGAGC | 21 |
| clfA | clfA-UpclfA-Dn | ATTGGCGTGGCTTCAGTGCTTGGCTTGATTGAGTTGTTGCCGGTGT | 21 |
| arcA | arcA-UparcA-Dn | CACGTAACTTGCTAGAACGAGGAGCCAGAAGTACGCGAG | 21 |
| icaA | icaA-UpicaA-Dn | TCAGACACTTGCTGGCGCAGTCTCACGATTCTCTCCCTCTCTGCCATT | 21 |
S. aureus isolates harbouring virulence genes determined in our previous study were used as positive control strains for detecting virulence genes.20
SCCmec typingSCCmec typing of MRSA isolates was performed using a battery of multiplex PCRs as described previously.23 MRSA isolates with unanticipated fragments or lacking fragments by multiplex PCR were defined as non-typeable (NT). MRSA NCTC 10442 (SCCmecI), MRSA N315 (SCCmec II), MRSA 85/2082 (SCCmec III), MRSA JCSC 4744 (SCCmec IV), and MRSA WZ153 (SCCmec V) were used as control strains for SCCmec typing.
spa typingThe spa variable repeat region from each S. aureus isolate was amplified using simplex PCR oligonucleotide primers as previously described.24,25 Following their purification and sequencing, spa types were assigned using the spa database website (http://www.ridom.de/spaserver).
Multi-locus sequence typing (MLST)MLST typing of S. aureus isolates was performed using amplification of internal fragments of the seven housekeeping genes of S. aureus as described previously.26 Following purification and sequencing of these genes, the sequences were compared with the existing sequences available on the MLST website for S. aureus (http://saureus.mlst.net), and STs were determined according to the allelic profiles. Novel STs were deposited in the MLST database (http://saureus.mlst.net/).
Statistical analysisDifferences between groups were assessed by using the chi-square test. The software SPSS 13.0 was used to perform calculations. p-Values of <0.05 were considered statistically significant.
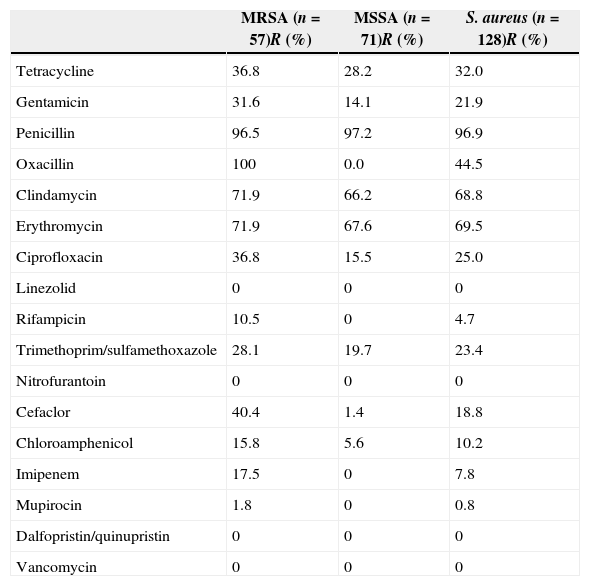
Results and discussionAntimicrobial susceptibilityAmong 128 S. aureus isolates, 57 (44.5%) were identified as MRSA determined by cefoxitin disc diffusion test and were positive for mecA. The MRSA prevalence in the present study was lower than the 54.1% reported between December 2002 and June 2008.11 The resistance rates for S. aureus, MRSA, and MSSA isolates to antimicrobials are listed in Table 2. All isolates tested were susceptible to vancomycin, linezolid, dalfopristin/quinupristin, and nitrofurantoin. Of 128 S. aureus isolates, 72.7% (93/128) with resistance to three or more classes of antimicrobial agents tested were defined as multidrug-resistant isolates. Only two isolates tested were susceptible to all antimicrobial agents tested. Twenty-three (18.0%) isolates were only resistant to penicillin. Ten (7.8%) isolates were concomitantly resistant to two antimicrobial agents tested (penicillin and another antimicrobial agent). Resistance rates of S. aureus, MRSA, and MSSA isolates to penicillin, clindamycin, and gentamicin were above 60%, whereas to tetracycline, gentamicin, ciprofloxacin, trimethoprim/sulfamethoxazole, chloramphenicol and rifampicin were less than 40%. Three isolates were positive for D-test, indicating that resistance of these isolates to clindamycin was inducible. Only one MRSA isolate was resistant to mupirocin as it exhibited no zone of inhibition.
Antimicrobial resistance profiles of MRSA, MSSA, and S. aureus isolates.
| MRSA (n=57)R (%) | MSSA (n=71)R (%) | S. aureus (n=128)R (%) | |
|---|---|---|---|
| Tetracycline | 36.8 | 28.2 | 32.0 |
| Gentamicin | 31.6 | 14.1 | 21.9 |
| Penicillin | 96.5 | 97.2 | 96.9 |
| Oxacillin | 100 | 0.0 | 44.5 |
| Clindamycin | 71.9 | 66.2 | 68.8 |
| Erythromycin | 71.9 | 67.6 | 69.5 |
| Ciprofloxacin | 36.8 | 15.5 | 25.0 |
| Linezolid | 0 | 0 | 0 |
| Rifampicin | 10.5 | 0 | 4.7 |
| Trimethoprim/sulfamethoxazole | 28.1 | 19.7 | 23.4 |
| Nitrofurantoin | 0 | 0 | 0 |
| Cefaclor | 40.4 | 1.4 | 18.8 |
| Chloroamphenicol | 15.8 | 5.6 | 10.2 |
| Imipenem | 17.5 | 0 | 7.8 |
| Mupirocin | 1.8 | 0 | 0.8 |
| Dalfopristin/quinupristin | 0 | 0 | 0 |
| Vancomycin | 0 | 0 | 0 |
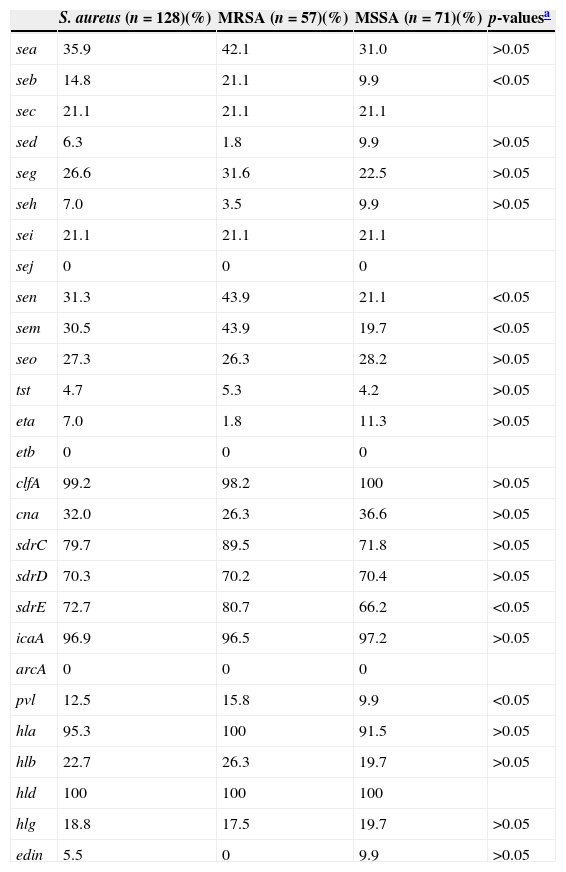
The invasiveness of S. aureus largely depends on the carriage of a battery of virulence factors.2,3 All S. aureus SSTI isolates in the present study harboured at least five virulence genes tested. Frequencies of virulence genes are shown in Table 3. Virulence genes were detected among at least 70% of all tested isolates included hld (100%), hla (95.3%), icaA (96.9%), clf (99.2%), sdrC (79.7%), sdrD (70.3%), and sdrE (72.7%). Less than 10% of the isolates tested carried eta (7.0%), sed (6.3%), seh (7.0%), tst (4.7%), and edin (5.5%). Multiple isolates harboured pvl (12.5%), sea (35.9%), seb (14.8%), sec (21.1%), sei (21.1%), seg (26.6%), sem (30.5%), sen (31.3%), seo (27.3%), hlb (22.7%), hlg (18.8%), and cna (32.0%). All S. aureus isolates tested were negative for sej, etb, and arcA. Fifty-six (43.75%, 56/128) isolates harboured more than 10 tested virulence genes, among which two isolates harboured 16 genes, seven isolates 14 genes, 13 isolates 13 genes, six isolates 12 genes, 10 isolates 11 genes, and 18 isolates 10 genes. Of MRSA isolates, 54.4% (31/56) harboured more than 10 tested virulence genes, which was significantly higher than that among MSSA isolates (35.2%, 25/71) (p<0.05).
The frequencies of virulence genes among S. aureus, MRSA and MSSA isolates.
| S. aureus (n=128)(%) | MRSA (n=57)(%) | MSSA (n=71)(%) | p-valuesa | |
|---|---|---|---|---|
| sea | 35.9 | 42.1 | 31.0 | >0.05 |
| seb | 14.8 | 21.1 | 9.9 | <0.05 |
| sec | 21.1 | 21.1 | 21.1 | |
| sed | 6.3 | 1.8 | 9.9 | >0.05 |
| seg | 26.6 | 31.6 | 22.5 | >0.05 |
| seh | 7.0 | 3.5 | 9.9 | >0.05 |
| sei | 21.1 | 21.1 | 21.1 | |
| sej | 0 | 0 | 0 | |
| sen | 31.3 | 43.9 | 21.1 | <0.05 |
| sem | 30.5 | 43.9 | 19.7 | <0.05 |
| seo | 27.3 | 26.3 | 28.2 | >0.05 |
| tst | 4.7 | 5.3 | 4.2 | >0.05 |
| eta | 7.0 | 1.8 | 11.3 | >0.05 |
| etb | 0 | 0 | 0 | |
| clfA | 99.2 | 98.2 | 100 | >0.05 |
| cna | 32.0 | 26.3 | 36.6 | >0.05 |
| sdrC | 79.7 | 89.5 | 71.8 | >0.05 |
| sdrD | 70.3 | 70.2 | 70.4 | >0.05 |
| sdrE | 72.7 | 80.7 | 66.2 | <0.05 |
| icaA | 96.9 | 96.5 | 97.2 | >0.05 |
| arcA | 0 | 0 | 0 | |
| pvl | 12.5 | 15.8 | 9.9 | <0.05 |
| hla | 95.3 | 100 | 91.5 | >0.05 |
| hlb | 22.7 | 26.3 | 19.7 | >0.05 |
| hld | 100 | 100 | 100 | |
| hlg | 18.8 | 17.5 | 19.7 | >0.05 |
| edin | 5.5 | 0 | 9.9 | >0.05 |
Staphylococcal enterotoxins (SEs), including five major classical antigenic types of SEs (SEA to SEE) and newly identified SEs are the cause of food poisoning in humans.27 Additionally, SEs are also associated with other diseases such as allergy sensitization, asthma, chronic obstructive pulmonary disease, scarlet fever, glomerulonephritis, and vasculitis.28–31 The genes encoding these SEs but sej were found among S. aureus SSTI isolates, with different carriage proportions ranging from 6.3% to 35.9% in this investigation. In particular, the positive rates for seb, sen, and sem among MRSA isolates were significantly higher than among MSSA isolates (p<0.05). The Sdr proteins encoded by the tandemly arrayed sdrC, sdrD, and sdrE are microbial surface components which recognize adhesive matrix molecules and have different roles in S. aureus pathogenicity.32 Strong correlations between S. aureus invasiveness and the presence of one of the allelic variants of the sdrE gene, as well as carriage of the sdrD gene and bone infections caused by S. aureus, have been reported previously.33,34 Our previous study showed that 95.5% (85/89) of S. aureus isolates responsible for bloodstream infection harboured at least one sdr locus and 84.3% (75/89) possessed more than two sdr loci.35 Similarly, 120 (93.8%) of 128 SSTI isolates in the present study were found to harbour at least one sdr locus (sdrC, sdrD, or sdrE), with 18 (14.1%), 39 (30.5%), and 63 (49.2%) harbouring one, two, or three of these loci, respectively. The positive rate of sdrE among MRSA isolates was significantly higher than among MSSA isolates. S. aureus isolates producing TSST-1 encoded by the tst gene have been associated with toxic shock syndrome, staphylococcal scarlet fever, and neonatal toxic shock-like exanthematous diseases.3 However, the present study found that only 4.7% of S. aureus SSTI isolates harboured tst. Our previous study found that the prevalence of hlb among S. aureus isolates associated with bloodstream infection was 67.4%.35 However, hlb was only identified among 22.7% of S. aureus SSTI isolates, while the positivity rates of hla and hld were 95.3% and 100%, respectively.
pvl has been closely associated with CA-MRSA infections and there is a strong epidemiological association between carriage of pvl genes and successful CA-MRSA lineages.1,36 Infections caused by pvl-positive S. aureus isolates are predominantly represented by skin and soft-tissue infection.5,6 Among S. aureus isolates causing SSTI in our hospital between December 2002 and June 2008, the overall positivity rates of pvl genes 23.4% (26/111), and among MRSA and MSSA isolates the rates were, 21.7% (13/60) and 25.5% (13/51), respectively.11 Compared with our previous study, the overall positivity rates of pvl genes in the present study (12.5%) was lower, as were the rates among MRSA (15.8%) and MSSA (9.9%) isolates, indicating that there is a decreased trend in the prevalence of pvl genes among S. aureus SSTI isolates at our hospital.
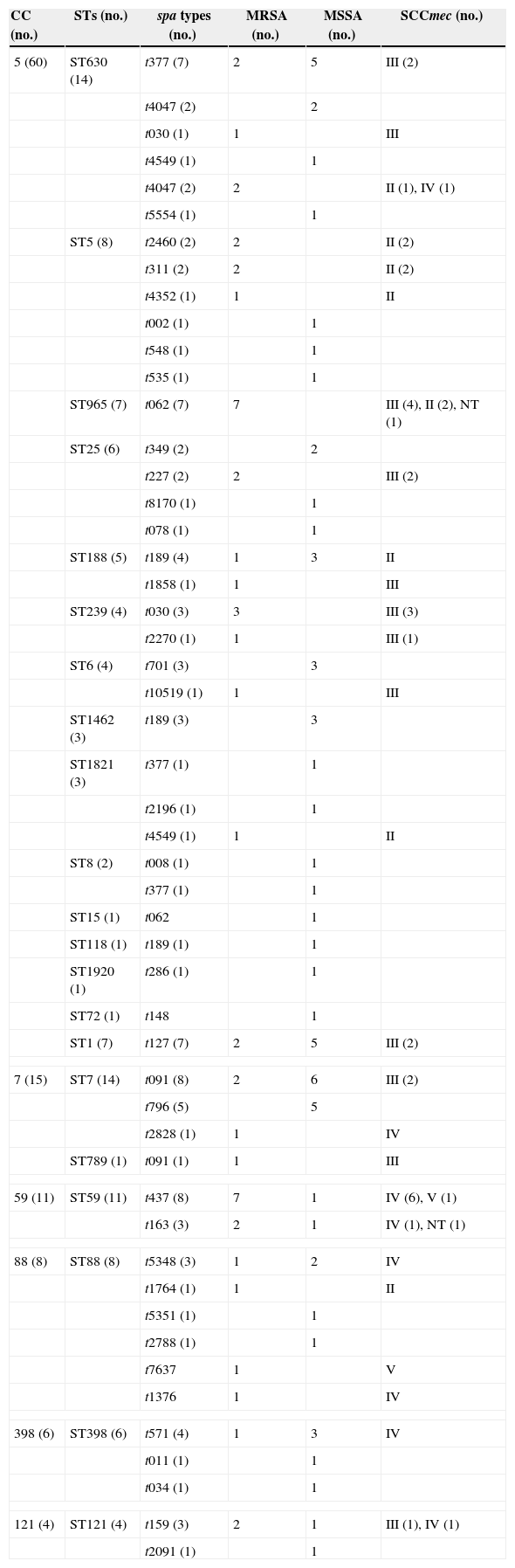
Molecular typingMolecular typing of S. aureus isolates tested are shown in Table 4. Among 57 MRSA isolates, 24, 14, 13, and three harboured SCCmec types III, IV, II, and V, respectively. Three isolates were classified as non-typeable.
Molecular characteristics of S. aureus SSTIs isolates.
| CC (no.) | STs (no.) | spa types (no.) | MRSA (no.) | MSSA (no.) | SCCmec (no.) |
|---|---|---|---|---|---|
| 5 (60) | ST630 (14) | t377 (7) | 2 | 5 | III (2) |
| t4047 (2) | 2 | ||||
| t030 (1) | 1 | III | |||
| t4549 (1) | 1 | ||||
| t4047 (2) | 2 | II (1), IV (1) | |||
| t5554 (1) | 1 | ||||
| ST5 (8) | t2460 (2) | 2 | II (2) | ||
| t311 (2) | 2 | II (2) | |||
| t4352 (1) | 1 | II | |||
| t002 (1) | 1 | ||||
| t548 (1) | 1 | ||||
| t535 (1) | 1 | ||||
| ST965 (7) | t062 (7) | 7 | III (4), II (2), NT (1) | ||
| ST25 (6) | t349 (2) | 2 | |||
| t227 (2) | 2 | III (2) | |||
| t8170 (1) | 1 | ||||
| t078 (1) | 1 | ||||
| ST188 (5) | t189 (4) | 1 | 3 | II | |
| t1858 (1) | 1 | III | |||
| ST239 (4) | t030 (3) | 3 | III (3) | ||
| t2270 (1) | 1 | III (1) | |||
| ST6 (4) | t701 (3) | 3 | |||
| t10519 (1) | 1 | III | |||
| ST1462 (3) | t189 (3) | 3 | |||
| ST1821 (3) | t377 (1) | 1 | |||
| t2196 (1) | 1 | ||||
| t4549 (1) | 1 | II | |||
| ST8 (2) | t008 (1) | 1 | |||
| t377 (1) | 1 | ||||
| ST15 (1) | t062 | 1 | |||
| ST118 (1) | t189 (1) | 1 | |||
| ST1920 (1) | t286 (1) | 1 | |||
| ST72 (1) | t148 | 1 | |||
| ST1 (7) | t127 (7) | 2 | 5 | III (2) | |
| 7 (15) | ST7 (14) | t091 (8) | 2 | 6 | III (2) |
| t796 (5) | 5 | ||||
| t2828 (1) | 1 | IV | |||
| ST789 (1) | t091 (1) | 1 | III | ||
| 59 (11) | ST59 (11) | t437 (8) | 7 | 1 | IV (6), V (1) |
| t163 (3) | 2 | 1 | IV (1), NT (1) | ||
| 88 (8) | ST88 (8) | t5348 (3) | 1 | 2 | IV |
| t1764 (1) | 1 | II | |||
| t5351 (1) | 1 | ||||
| t2788 (1) | 1 | ||||
| t7637 | 1 | V | |||
| t1376 | 1 | IV | |||
| 398 (6) | ST398 (6) | t571 (4) | 1 | 3 | IV |
| t011 (1) | 1 | ||||
| t034 (1) | 1 | ||||
| 121 (4) | ST121 (4) | t159 (3) | 2 | 1 | III (1), IV (1) |
| t2091 (1) | 1 | ||||
A total of 28 STs were identified among 128 S. aureus isolates. ST7 and ST630 accounting for 10.9% (14/128 each) were found to be the predominant STs, followed by ST59 (8.6%, 11/128), ST5 (6.3%, 8/128), ST88 (6.3%, 8/128), ST1 (5.5%, 7/128), ST965 (5.5%, 7/128), ST398 (4.7%, 6/128 each), ST25 (4.7%, 6/128), and ST188 (3.9%, 5/128). ST239, ST6, and ST121 accounted for four isolates each. ST1463 and ST1821 accounted for three isolates each. ST8 and ST1349 accounted for two isolates each. The remaining STs including ST12, ST15, ST118, ST692, ST789, ST1281, ST1920, ST2259, ST2832, ST2833, and ST72 were identified in only one isolate. The STs of nine isolates were not identified. Two novel STs characterized as ST2832 and 2833 were identified in two MRSA isolates and have been deposited in the MLST database (http://saureus.mlst.net/). Sixteen PVL-positive isolates were distributed among nine different STs including ST88 (five isolates), ST59 (three isolates), and ST121 (two isolates). ST239 and ST5 were the most dominant STs in China.9,37 Interestingly, in the present study, these two predominant STs were found to be minor clones, while ST630 and ST7, seldom noted in Chinese isolates previously, were the major clones among S. aureus SSTI isolates. Another study from China found ST398 accounting for 17.1% (28/164) as the most common ST among S. aureus SSTI isolates.38 In contrast to our previous study where ST239 was the most prevalent ST accounting for 21.6% (24/111) of S. aureus SSTI isolates,11 this ST accounted for only 3.1% (4/128) in the present study. Interestingly, ST630 not found in our previous study turned out to be the predominant ST in the present study, while ST1018, the second most prevalent ST in our previous study, was not found.
Fifty-two spa types were identified among the 128 isolates. The most prevalent spa type was t091 (8.6%, 11/128), followed by t062 (7.0%, 9/128), t377 (7.0%, 9/128), t437 (7.0%, 9/128), t189 (6.3%, 8/128), t127 (4.7%, 6/128), t796 (3.9%, 4/128), t571 (3.1%, 3/128), t4047 (3.1%, 3/128), and t030 (3.1%, 3/128). Other types identified included t159, t163, t5348, and t701 (three isolates each). In previous reports of Chinese isolates, t30 and t37, typically associated with ST239, were the most prevalent spa types.9,15 The proportions of t37 and t30 were extremely low, as well as the proportions of ST239 and ST5 in the present study.
Fifty-seven MRSA isolates were distributed in different clones. 12.3% (7/57) of MRSA isolates belonged to MRSA-ST59-SCCmec IV-t 437/163, which was the most common clone in the present study. Likewise, in a study from China ST59-MRSA-IVa-t437 was found to be the predominant clone among CA-MRSA isolates associated with SSTIs in children.39 Our previous study conducted between December 2002 and June 2008 found that ST239-MRSA-SCCmec III accounting for 30.2% (19/63) was the most prevalent clone among MRSA SSTI isolates, followed by ST1018-MRSA-SCCmecIII accounting for 15.9% (10/63).11 However, only three MRSA isolates were ST239-MRSA-SCCmec III and there was no ST1018-MRSA-SCCmecIII isolate in the present study. Among 71 MSSA isolates, the dominant clone was MSSA-ST7 (15.5%, 11/71), followed by MSSA-ST630 (12.7%, 9/71), MSSA-ST398 and MSSA-ST1 (7.0%, 5/71 each), and MSSA-ST25 and MSSA-ST88 (5.6%, 4/71 each). MRSA-ST398 isolates are usually associated with infections in both animals and humans.40,41 ST398 MSSA isolates of human origin are usually linked to t571.42 In the present study, four of six ST-398 S. aureus isolates, including three MSSA and one MRSA isolate, were linked to t571.
Taken together, our data showed that S. aureus isolates associated with SSTIs were genetically diverse and the main clones associated with SSTIs are going through a rapid shift in our hospital.
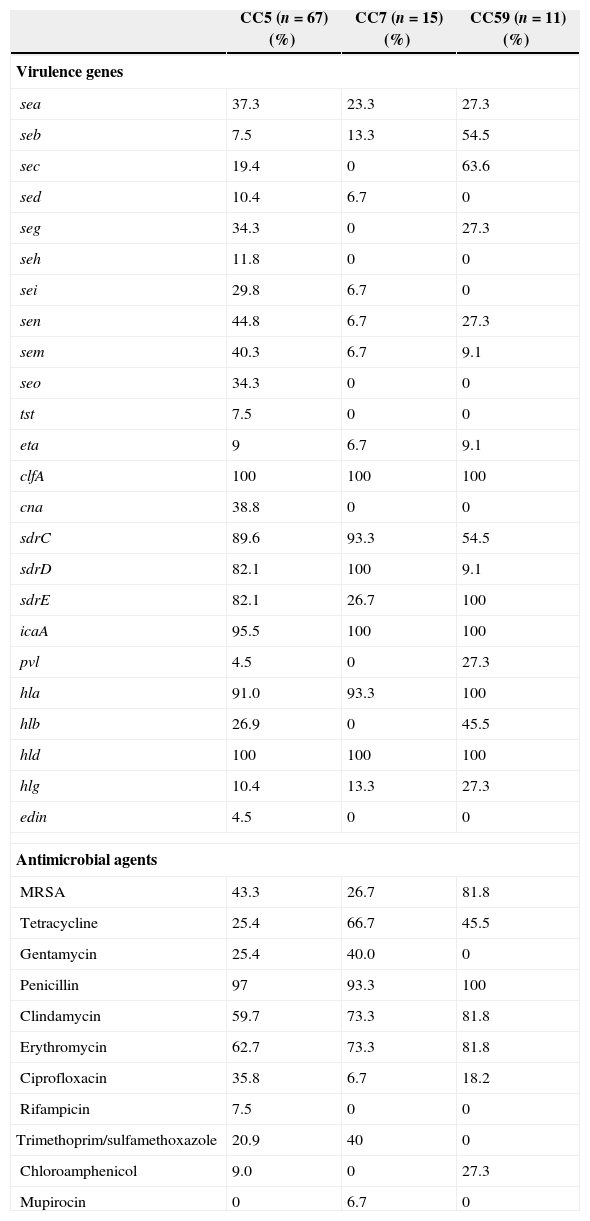
Comparison of antimicrobial resistance and molecular typing among the major clonal complexes (CCs)Clustering analysis by eBURST v3 showed that six clonal complexes (CCs) were found (Table 4), including CC5 (52.3%, 67/128), CC7 (11.7%, 15/128), CC59 (8.6%, 11/128), CC88 (6.3%, 8/128), and CC398 (4.7%, 6/128). The distribution of some virulence genes, especially enterotoxin genes, were correlated with different MRSA lineages.43,44 In the present study, a higher carriage of seb and sec was found among CC59 isolates, while a higher carriage of enterotoxin genes were found among CC5 isolates (Table 5). All CC7 and CC59 isolates were not carrying seh and seo, while 11.8% and 34.2% of CC5 isolates were found to carry these two genes, respectively. Although the proportions of seb and sec among CC5 and CC7 isolates were low, the positivity rates of these two enterotoxin genes among CC59 isolates were 54.5% (seb) and 63.6% (sec). The prevalence of cna among CC5 isolates was 38.8%, while this virulence gene was identified in none of the CC7 and CC59 isolates. Compared with the higher carriage rates (>80.0%) of sdrC and sdrD among CC5 and CC7 isolates, the positivity rates of two sdr loci were significantly lower among CC59 isolates, especially sdrD prevalence of 9.1%. Interestingly, all CC59 isolates carried sdrE, while only 26.7% of CC7 isolates were found to carry sdrE. The carriage rates of pvl and hlb among CC59 isolates were significantly higher than those rates among CC5 and CC7 isolates (p<0.05). These differences in carriage rates of virulence genes among different CC isolates suggested that different S. aureus lineages associated with SSTIs have specific patterns of virulence genes.
Specific antimicrobial resistance and virulence gene profiles of major CCs.
| CC5 (n=67) (%) | CC7 (n=15) (%) | CC59 (n=11) (%) | |
|---|---|---|---|
| Virulence genes | |||
| sea | 37.3 | 23.3 | 27.3 |
| seb | 7.5 | 13.3 | 54.5 |
| sec | 19.4 | 0 | 63.6 |
| sed | 10.4 | 6.7 | 0 |
| seg | 34.3 | 0 | 27.3 |
| seh | 11.8 | 0 | 0 |
| sei | 29.8 | 6.7 | 0 |
| sen | 44.8 | 6.7 | 27.3 |
| sem | 40.3 | 6.7 | 9.1 |
| seo | 34.3 | 0 | 0 |
| tst | 7.5 | 0 | 0 |
| eta | 9 | 6.7 | 9.1 |
| clfA | 100 | 100 | 100 |
| cna | 38.8 | 0 | 0 |
| sdrC | 89.6 | 93.3 | 54.5 |
| sdrD | 82.1 | 100 | 9.1 |
| sdrE | 82.1 | 26.7 | 100 |
| icaA | 95.5 | 100 | 100 |
| pvl | 4.5 | 0 | 27.3 |
| hla | 91.0 | 93.3 | 100 |
| hlb | 26.9 | 0 | 45.5 |
| hld | 100 | 100 | 100 |
| hlg | 10.4 | 13.3 | 27.3 |
| edin | 4.5 | 0 | 0 |
| Antimicrobial agents | |||
| MRSA | 43.3 | 26.7 | 81.8 |
| Tetracycline | 25.4 | 66.7 | 45.5 |
| Gentamycin | 25.4 | 40.0 | 0 |
| Penicillin | 97 | 93.3 | 100 |
| Clindamycin | 59.7 | 73.3 | 81.8 |
| Erythromycin | 62.7 | 73.3 | 81.8 |
| Ciprofloxacin | 35.8 | 6.7 | 18.2 |
| Rifampicin | 7.5 | 0 | 0 |
| Trimethoprim/sulfamethoxazole | 20.9 | 40 | 0 |
| Chloroamphenicol | 9.0 | 0 | 27.3 |
| Mupirocin | 0 | 6.7 | 0 |
The predominant MRSA clones in China were associated with specific antimicrobial resistance profiles.45 In the present study, although 81.8% of CC59 isolates were MRSA, all isolates were susceptible to gentamicin, rifampicin, and trimethoprim/sulfamethoxazole (Table 5). However, the resistance rates for gentamicin and trimethoprim/sulfamethoxazole were respectively 25.4% and 20.9% among CC5 isolates, and 40.0% and 40.0% among CC7 isolates (Table 5). The resistance rates for tetracycline, clindamycin, and erythromycin among CC5 isolates were relatively lower relative to CC7 and CC59 isolates (Table 5). As the prevalence of MRSA among CC7 isolates was lower than CC5 and CC59 isolates, the resistance rates of some antimicrobial agents among CC7 isolates were lower than other CC isolates, such as ciprofloxacin and chloramphenicol. However, only one isolate with resistance to mupirocin in this investigation belonged to CC7.
In conclusion, the molecular characteristic of S. aureus SSTI isolates in the present study showed considerable heterogeneity and ST7 and ST630 were the prevalent clones. Different S. aureus clones causing SSTIs were associated with specific antimicrobial resistance and virulence gene profiles.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by grants from Natural Science Fund of China (81271906H2002), Medical Science Fund of Zhejiang Province, China (2012RCA041) and Wenzhou Municipal Science and Technology Bureau, China (Y20130239).