Toxoplasmosis is a zoonotic infection caused by the protozoan parasite Toxoplasma gondii. The infection is widely disseminated in the human population and is usually benign or asymptomatic. Systemic T. gondii infection presents risks for pregnant women and AIDS patients. Although rare, T. gondii can cause outbreaks in urban centers. The origin of these outbreaks is not completely understood but probably results from introduction of zoonotic T. gondii strains in the population. During such outbreaks other pathogens which mimic T. gondii acute febrile syndrome may also circulate; therefore, detailed investigation of the outbreak is of extreme importance. In this study we performed viral metagenomics next-generation sequencing (mNGS) in patient samples obtained during T. gondii outbreak in Santa Maria city, South Brazil. Specific bioinformatics pipelines specialized in virus discovery were applied in order to identify co-circulating vial agents. Epstein Barr virus and Parvovirus B19 contigs were assembled and these viruses can cause symptoms similar to toxoplasmosis. In conclusion, our findings show the importance of Metagenomics next generation sequencing (mNGS) use to help characterize the outbreak more completely and in the management of the affected patients.
Toxoplasma gondii is a protozoan parasite that causes toxoplasmosis, a worldwide spread zoonotic infection, which is also highly prevalent in humans. The infection, in most cases is asymptomatic or has benign clinical course. However, T. gondii can cause life-threatening conditions when it is transmitted congenitally from mother to fetus or when the parasite infects patients with immune deficiency, especially AIDS.1
After birth, human toxoplasmosis is acquired by ingesting water or undercooked meat contaminated with oocysts.2–4 For that reason, sporadic outbreaks of T. gondii can occur, and are concentrated in small groups of individuals who have consumed contaminated food or inhaled oocysts from animal feces.5–8 In Brazil, T. gondii outbreaks have been reported in different regions of the country (Northeast, Southeast, South), both in groups of individuals or urban areas.9–11 The investigation of the origin of a T. gondii outbreak is difficult mainly due to the similarity of the symptoms with other infectious agents, especially viruses, the circulation of wild T. gondii strains, and the wide dissemination of this infection. Toward that end, a suitable method for identifying circulating infectious agents during an outbreak is the application of metagenomics next generation sequencing (mNGS), which is an unbiased technique providing information on the genomic composition of clinical samples and consequently the etiologic nature of the outbreak.
In April 2018, the largest T. gondii outbreak in Brazil was registered in the city of Santa Maria, Rio Grande do Sul State, South Brazil. By this period, there was a significant increase in patients who represented symptoms like fever, myalgia, headache, lymphadenopathy, and acute febrile syndrome (mononucleosis like disease). Until, June, 2018 there were 510 confirmed cases of toxoplasmosis in this location in almost all residential areas, as reported by the Municipal Sanitary Agency (report available at http://www.santamaria.rs.gov.br/docs/noticia/2018/06/D08-1465.pdf). In order to observe if viral agents also circulated during that outbreak, we performed a viral metagenomic survey aiming to investigate possible viral causative agents for the observed symptoms.
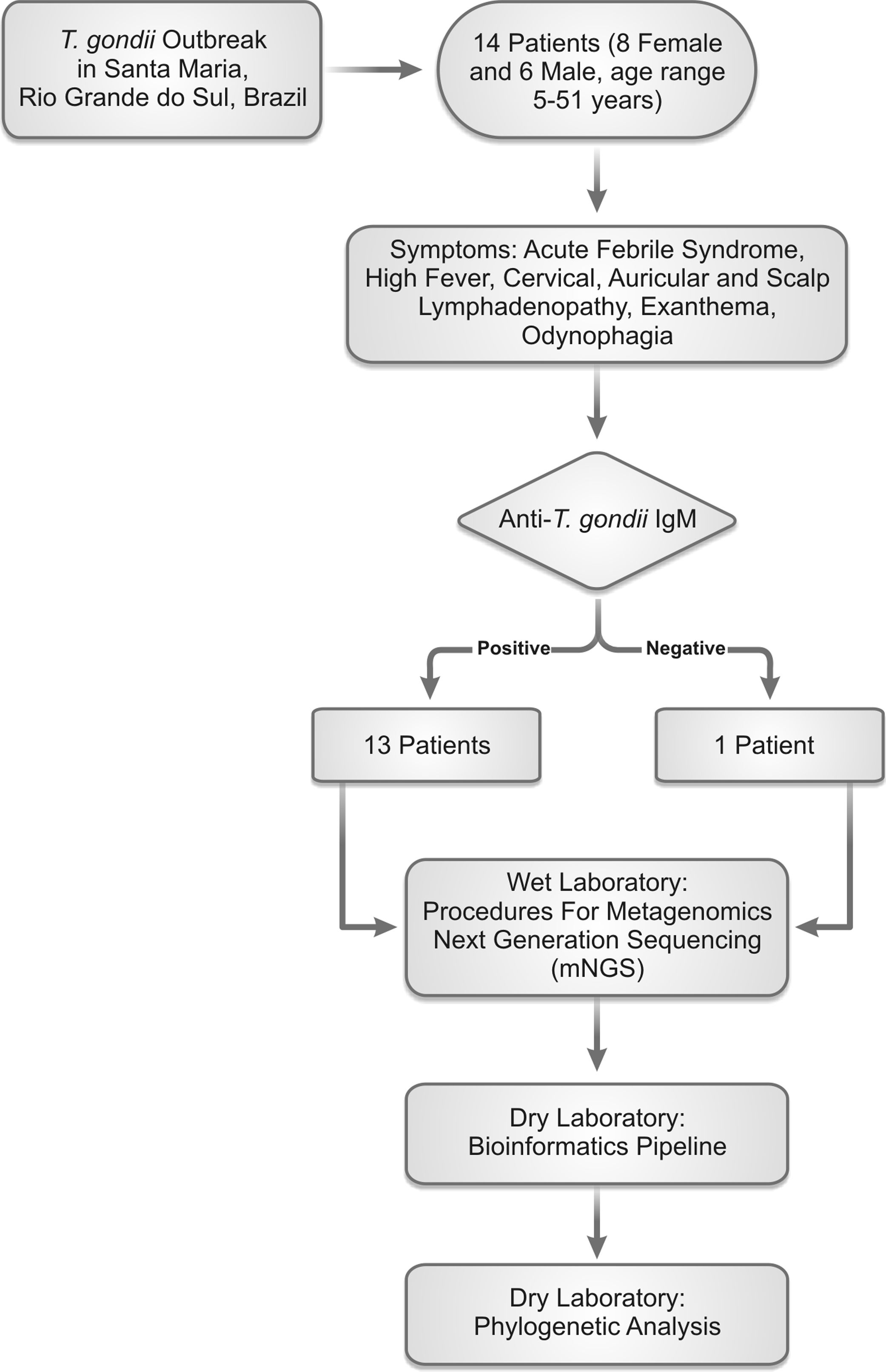
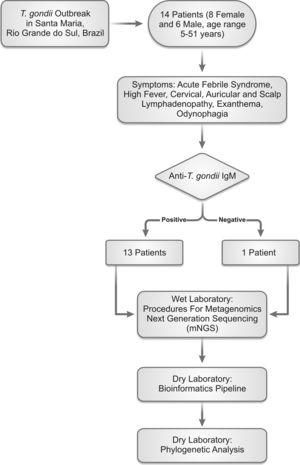
Materials and methodsA total of 14 plasma samples collected from patients testing positive for anti-T. gondii IgM and IgG (except one) during the outbreak were collected. All patients presented symptoms of acute febrile systemic disease. On average, the samples were collected two weeks after appearance of the acute symptoms. Peripheral blood samples were collected by brachial vein puncture from each patient using sterile EDTA K3 tubes (INJEX VÁCUO, Ourinhos, São Paulo, Brazil). For plasma separation from blood cell components, the samples were centrifuged at low speed (1455×g, 10min). All of the patients were individually informed about the study and signed a written informed consent. Santa Maria city is a medium-sized city, located in the central part of the Rio Grande do Sul State (29° 41’ 02” S 53° 48’ 25” W).
The mean age of the patients was 31.85 years (range 5–51 years), 57.1% were female and 42.9% male. All patients had a clinical condition compatible with systemic toxoplasmosis, which included myalgia, asthenia, prolonged high fever, and lymphadenopathy. No ocular manifestations were reported. One of the patients was pregnant and presented spontaneous abortion.
Before nucleic acid extraction, 800μL of each sample was centrifuged (17,000×g, 5min) for removal of plasma proteins and cell debris. Consequently, a volume of 600μL of plasma was separated and treated with Turbo DNase (Turbo DNA-free kit, ThermoFisher Scientific, São Paulo, Brazil) to remove free host/bacterial/protozoan DNA (incubation at 37°C for 30min). After DNAse inactivation, all 14 samples were mixed in order to form a sample pool. The total volume of the pool was used to extract DNA using the High Pure Viral Nucleic Acid Large Volume Kit (Roche, São Paulo, Brazil) following the manufacturer's instructions. Reverse transcription was performed using Superscript III First-Strand Synthesis System (ThermoFisher Scientific, São Paulo Brazil) and amplification of cDNA was carried out by the QuantiTect Whole Transcriptome Kit (QIAGEN, São Paulo, Brazil). The library preparation was performed using the Nextera DNA Flex Library Preparation Kit (Illumina, San Diego, CA, USA) and the Nextera DNA CD Indexes kit following the manufacturer's instructions. The sequencing of the libraries was carried out in Illumina NextSeq 500 sequencer using the NextSeq High Output Kit v 2.5, 300 cycles (Illumina, San Diego, CA, USA) following manufacturer's instructions. To control the mNGS reactions we used the PhiX Sequencing Control v.3 (Illumina, San Diego, CA, USA); however, due to the high cost of the sequencing internal positive and/or blank controls were not used.
The raw sequence data was subjected to quality control analysis using FastQC v.0.11.8 software. After evaluation the quality of the obtained reads, trimming was performed for selection of the sequences with best quality. Trimming was done in two distinct ways: (i) by the use of Trimmomatic v.0.3.9 for removing low quality sequences, duplicates and ambiguous bases; and (ii) Cutadapt v. 2.4 for removal of Illumina adapters. For subsequent analysis we used only sequences with an overall quality of above 30 scores. Additionally, we performed a second trimming step using the AfterQC v.0.9.7 software to remove poliA tails and more ambiguous bases.
A bioinformatic pipeline was developed focusing on viral metagenomics analysis, but we were also able to identify some protozoa because our custom database contained protozoan taxonomic information. To infer the taxonomic classification, we used Kraken2 program. The “de novo” assembly was performed using SPAdes v.3.13.0. Finally, in order to identify the nucleotide and amino acid similarity we performed a Blastn and after a Blastx approach useing Diamond v 0.9.29.
The phylogenetic analysis was performed using a dataset of 45 complete genome sequences for Parvovirus B19 (B19V) and 20 complete genomes from Epstein Barr virus (EBV).
The alignment was performed using MAFFT v.7.429 and the phylogenetic signal was evaluated the Tree Puzzle v.5.2 program. A phylogenetic tree was built with IQtree v.16.12 applying the maximum likelihood algorithm with statistic support of ultrafast bootstrap with 10,000 replicates. The nucleotide substitution model was TN+F+R2 for B19V and K3Pu+F+R2 for EBV chosen according to BIC (Bayesian Information Criterion) statistic model. The final formatting and visualization of the phylogenetic tree was performed using the FigTree v.1.4.4 software.
Finally, we performed direct confirmation of the presence of B19V DNA in the plasma samples present in the tested pool. For B19V DNA detection TaqMan® real-time PCR was used with primers and probes already described in the literature.12 The amplification was performed in Applied Biosystems® 7500 real-time PCR system and all measures to prevent contamination were strictly adhered to.
ResultsThe performed bioinformatic analysis on the obtained samples, revealed in first place the presence of reads, which were taxonomically classified as T. gondii (total number of 240 obtained from the whole pool). However, after Blastn screening, only the most representative sequences were considered for analysis (88 reads with high genetic similarity for T. gondii). Despite efforts to better characterize the infecting T. gondii strain, we were not able to perform the assembly of a larger contig. This was probably due to the method applied for DNA extraction, that includes treatment with high DNAse concentration, which may have degraded T. gondii genome. Moreover, we worked with plasma samples possibly with low parasitemia since T. gondii is an obligatory intracellular parasite. The BLASTn analysis demonstrated that the infecting T. gondii strain was classified as VEG strain (type III); however, one must take into account the small size of the analyzed sequences (average size of 150bp) which may generate inaccurate classification compared to robust phylogenetic approaches.
Regarding the viral abundance of the analyzed samples, human commensal viruses like Human Pegivirus-1 (HPgV-1) and Torque teno virus (TTV) types were identified. This is a normal finding as these viruses are highly prevalent in the human population. The identified HPgV-1 belonged to genotype 2 (Subgenotype 2A). From the TTV were able to identify the types TTV-11, 19 and 20.
Our analysis was also able to detect known viral pathogens which were circulating concomitantly with T. gondii during the outbreak and could be responsible for a clinical picture similar to toxoplasmosis. In the tested pools we were able to assemble half complete genome of B19V (2619bp) and smaller contig (381bp) belonging to EBV.
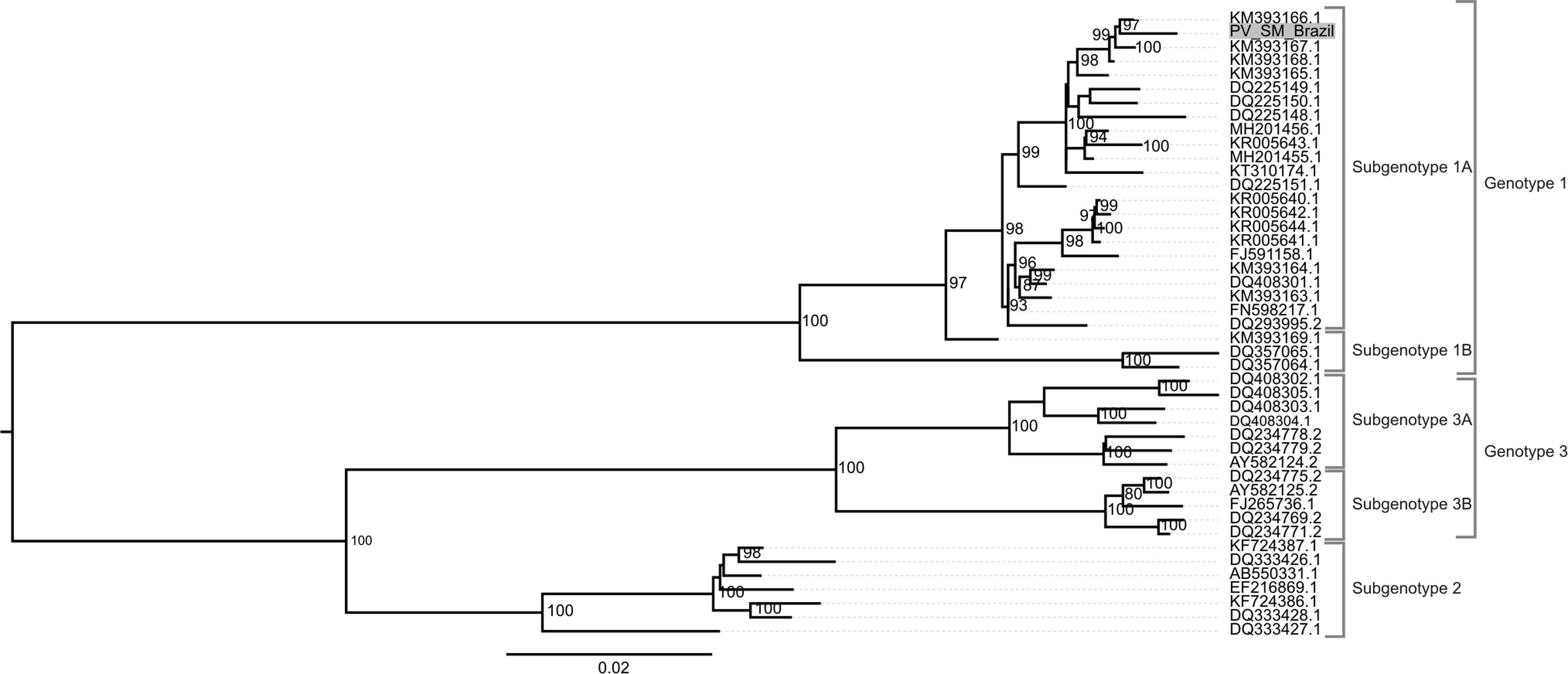
Phylogenetic classification was performed on the contigs of both viruses. B19V taxonomy was carried out using the maximum likelihood method with statistic support of ultrafast bootstrap with 10,000 replicates. The generated phylogenetic tree demonstrated the expected for B19V topology, with division of the strains into three genotypes. The identifed B19V strain (GenBank acession number MN556782) was characterized as belonging to subgenotype 1A of the B19V genotype 1. This strain was grouped with the KU5 isolate which has circulated in the USA during 2013 (for more information on the B19V observe Fig. 1).
Phylogenetic analysis of Parvovirus B19 (B19V). Only complete genomes were used for tracing the phylogenetic history of B19V. The nucleotide substitution model used was TN+F+R2 for tree reconstruction, which was chosen by BIC (Bayesian Information Criterion) statistic model, utilizing 10,000 ultrafast bootstrap replicates for statistical significance. Only values of above 75% were demonstrated on important tree branches. The phylogenetic tree was constructed using the IQtree software v.16.12, applying the maximum likelihood approach.
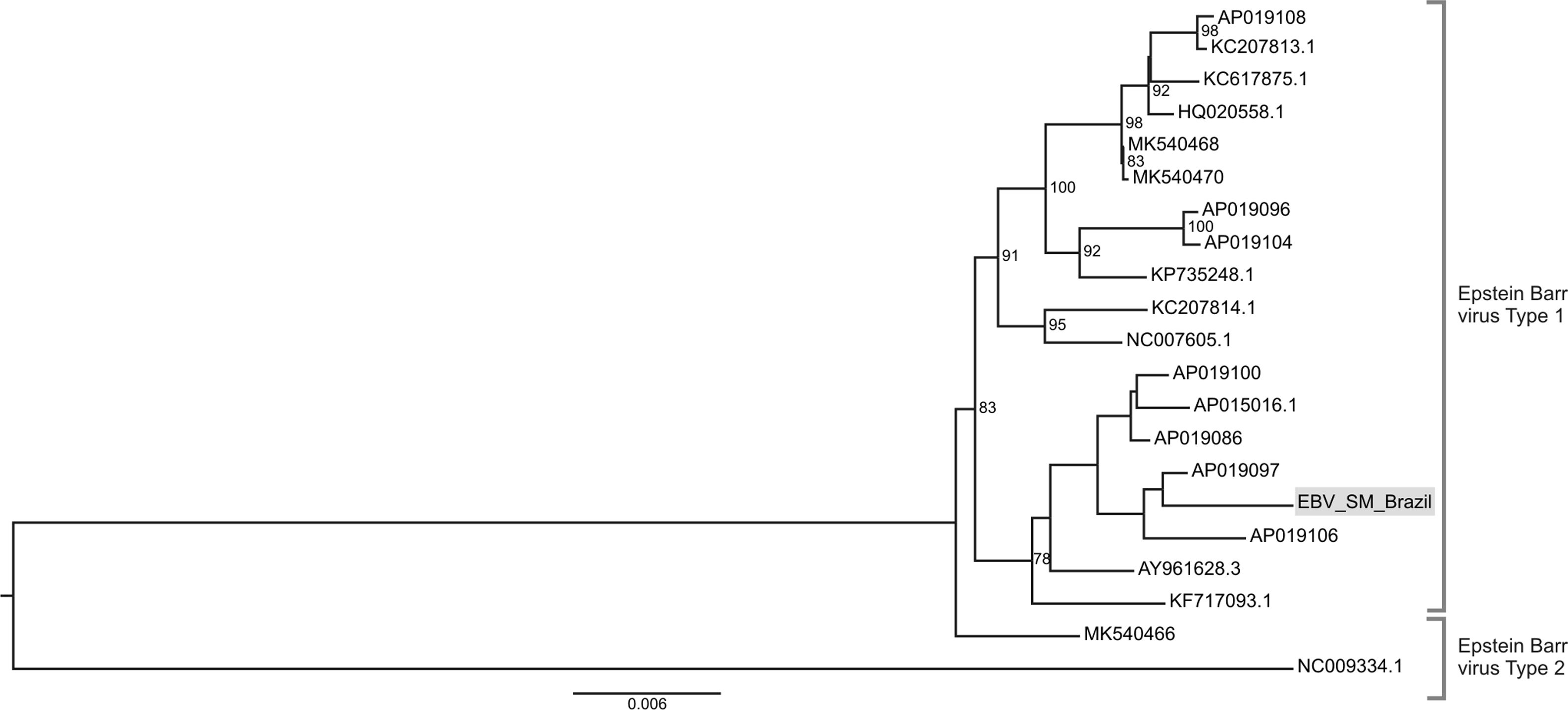
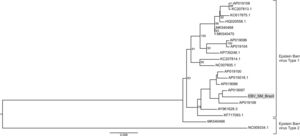
We also perfomed a phylogenetic analysis of the obtained EBV contig deposited as GenBank accession number MN551095.. The presence of EBV DNA sequences is justified by the finding that four of the positive for T. gondii showed also anti-EBV IgM seroreactivity. The performed EBV phylogenetic analysis demonstrated that our sequence belongs to the BBRF1 region of EBV genome, responsible for capsid portal protein synthesis (positions 101,916bp and ends at 103,757bp). The location of the detected sequence was between nucleotide positions 103,040bp and 103,338bp within the reference genome Human gammaherpesvirus 4 isolate UPN252 (AP019097). The phylogenetic analysis demonstrated the division of the evolutionary tree into the two classic EBV types with our strain belonging to EBV type 1 (Fig. 2). The sequence data from the NGS was deposited in the GenBank under the following number SRR7245009.
Phylogenetic analysis of Epstein Barr virus (EBV). Only complete genomes were used for phylogenetics analysis. The nucleotide substitution model used was K3Pu+F+R2 for tree reconstruction and was chosen according to BIC (Bayesian Information Criterion) statistic model, utilizing 10,000 ultrafast bootstrap replicates for statistical significance. Only values of above 75% are demonstrated on the important tree branches. The phylogenetic tree was constructed using the IQtree software v.16.12, applying the maximum likelihood approach.
The direct confirmation of B19V DNA demonstrated a positive result in one of the samples of a 35-year old male patient who presented acute febrile syndrome characterized by high fever and intense asthenia. The reaction cycle threshold (Ct) was 30.5 which probably was related to a viral load decrease. A limitation of our study was that we were not able to confirm the presence of EBV DNA by molecular methods. Nonetheless, we observed that four patients whose samples were included in the tested pool had positive results for anti-EBV IgM with different optical density (range 1.11 and 12U, reference values: 0.5U, not reactive; 0.5–0.99 borderline and >1.0 positive). Although the optical density is an indirect measure for confirmation of acute infection, further molecular testing is warranted in order to directly show the presence of viral DNA. The workflow of the performed study is shown in Fig. 3.
DiscussionIn this study we analyzed the virome of plasma samples from patients who presented acute febrile syndrome (mononucleosis like disease) during a T. gondii outbreak in South Brazil. The similarities of the clinical symptoms of toxoplasmosis to acute viral diseases warrant application of mNGS in order to identify other infectious agents which might co-circulate during the outbreak. In this respect we were able to identify co-circulating B19V and EBV, both causing acute febrile disease.
The performed metagenomic analysis detected the presence of sequence reads compatible with T. gondii VEG strain. Almost all patients presented anti-T. gondii IgM and IgG, which can be regarded as systemic toxoplasmosis.7T. gondii outbreaks have been described in different Brazilian areas including North, Southeast, and South regions.9,10,13 We were not able to determine the origin of the T. gondii strain due to the low number of specific T. gondii reads obtained during the mNGS. However, this could be an atypical strain of wild T. gondii which has entered the urban population. Such introductions of atypical T. gondii strains have been found in French Guiana and different Brazilian regions.9,14,15 This is reinforced by the fact that in South America, atypical T. gondii genotypes have been shown to circulate and can be responsible for the emergence of outbreaks with severe clinical presentation.16
We identified possible circulation of well-described pathogenic viruses during this outbreak. Larger contigs of B19V and EBV were assembled and primary infections with these agents might result in acute febrile disease. EBV, which causes infectious mononucleosis can mimic acute toxoplasmosis, and the differentiation based only on clinical findings is quite difficult.17 Therefore, application of mNGS can lead to identification of the respective pathogens and adoption of the most adequate treatment and management of the infection. Simultaneous presence of T. gondii and EBV has been described in a cord-blood transplant recipient leading to severe encephalitis18 and, therefore, establishing the most adequate treatment options has a crucial importance for patient management. Four of the patients presented anti-EBV IgM justifying detection by mNGS reads. The phylogenetic analysis demonstrated that the detected EBV strain belongs to type 1, which is typical in the Americas.19 We were also able to identify contigs belonging to B19V and frequent circulation of this virus has been described in South Brazil20 which explains our findings. The phylogenetic analysis demonstrated that the infecting B19V was of genotype 1 (sub-genotype 1A) which is the most frequent genotype circulating in Brazil.20 The presence of B19V DNA was directly confirmed in one of the individual samples (Ct=30.5) showing the clinical utility of mNGS for the diagnosis of clinically important infections. The presence of B19V and T. gondii co-infection has unknown clinical outcome; however, fatal myocardial coinfections have been observed in patients with HIV.21
The applied mNGS methods showed co-circulation of viral agents in individuals with acute febrile syndrome during T. gondii outbreak and, therefore, mNGS methods should be implemented to unravel the etiology of outbreaks of unknown origin. As mNGS is an unbiased strategy for pathogen discovery, it can bring important information of how to manage the outbreak, detect the genetic diversity of the circulating strains (origin of the outbreak), prevention of their dissemination, and importantly, treatment options depending on the circulating pathogen. For that reason, mNGS has already been successfully applied for investigating outbreaks of unknown etiology.22,23 We also believe that mNGS must be widely recommended as a first line tool for testing patient samples when deciding on the cause of a given outbreak and its subsequent management.
In conclusion, the performed study examined the importance of mNGS for identifying viral infections co-circulating during an outbreak of toxoplasmic acute febrile disease in South Brazil. As shortcoming of this study, we were not able to directly confirm all identified viral pathogens in the clinical samples; nevertheless, the performed mNGS revealed that patient's virome may be used as a viral diagnostic marker. The performance of metagenomic studies has several limitations including the high cost of the reaction which hampers testing larger numbers of individual samples and performing population-based studies. Nevertheless, the conducted study demonstrates the utility of mNGS not only for viral detection, but also for policy making decisions on how to manage the outbreak and consequently the infected patients.
FundingThe financial support of this study was provided by São Paulo Research Foundation (FAPESP), grants numbers: 17/23205-8, 18/15826-5, 19/08528-0, 19/07861-8, INCTC-FAPESP 2014/50947 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grants numbers 465539/2014-9 and 455503/2014-1.
Ethical approvalThis study was approved by the Institutional Ethics Committee of the University Hospital of the Faculty of Medicine of Ribeirão Preto, University of São Paulo
Informed consentVerbal informed consent was obtained from all patients participating in this study.
Conflicts of interestThe authors declare no conflicts of interest.
We are grateful to all patients who donated their samples for the realization of this study. We are also grateful to Roberta Maraninchi Silveira and Kamila Chagas Peronni for the sample preparation. We acknowledge Sandra Navarro Bresciani for the artwork.