The genus Enterovirus, a member of the Picornavirus family, are RNA viruses that can cause poliomyelitis, hand-food-mouth disease, viral meningitis or meningoencephalitis, viral myocarditis and so on. MicroRNAs are a class of highly conserved, small noncoding RNAs recognized as important regulators of gene expression. Recent studies found that MicroRNAs play a significant role in the infection of Enterovirus, such as enterovirus 71, coxsackievirus B3 and other Enterovirus. Enteroviral infection can alter the expression of cellular MicroRNAs, and cellular MicroRNAs can modulate viral pathogenesis and replication by regulating the expression level of viral or host's genes. Herein, this review summarizes the role of MicroRNAs in enteroviral infection.
MiRNAs are a class of highly conserved, small noncoding RNAs molecules, which contain 18- to 25- nucleotides (nt) in almost all eukaryotes. MiRNAs regulate gene expression at the post-transcriptional level, influence important physiological and pathological processes.1 For example, miR-375 can promote beta-pancreatic differentiation in human induced pluripotent stem cells.2 In some tumor diseases, miRNAs hold a pivotal regulatory role. MiR-15a and miR-16 can induce the development of multiple myeloma.3 MiRNAs act on mRNAs by either translation inhibition or mRNA degradation, depending on the extent of complementarity between target mRNA and miRNA sequence. There are two main mechanisms: imperfect sequence complementarity between miRNA and mRNA results in translation inhibition,4,5 and perfect sequence complementarity leads to mRNA cleavage.6 The mechanism may be influenced by parameters such as the degree of sequence homology,1 target site multiplicity,7 the free energy of binding, target site secondary structure,8 and location of mRNA.9 MiRNAs can down-regulate the expression of their target mRNAs predominantly by binding to 3′ untranslated region (3′ UTR) of 28 mRNA.9,10 Recent evidences showed that miRNA can also bind to the 5′ UTR11 or coding regions12 of the target gene to regulate gene expression. Moreover, one miRNA can target multiple mRNAs and one mRNA can be controlled by multiple miRNAs.9,10,13 In conclusion, miRNAs play an important role in cell development, apoptosis, proliferation, and signal transduction.14 Additionally, fat and cholesterol metabolism, nerve development, hormone secretion, and immune response may also be affected by miRNAs.15,16
The genus Enterovirus (EV), one group of the picornavirus family, has the following species according to the International Committee on Taxonomy of Viruses: EV-A, which contains EV71 and several Coxsackievirus group A (CVA) viruses; EV-B (Coxsackievirus group B (CVB) viruses, CVA9, echoviruses and few other EV); EV-C (polioviruses (PV) 1–3, several CVA and few other EV); EV-D (EV-D68, EV-D111, EV 70 and EV74); EV-E, EV-F, EV-G, EV-H, EV-J, and the rhinoviruses (HRV-A, HRV-B, HRV-C). The polyprotein of EV consists of P1, P2, and P3 regions, which encode structural proteins (VP4-VP1) and non-structural proteins (2A, 2B, 2C and 3A, 3B, 3C, 3D) respectively. EV infected millions of people worldwide in recent years, resulting in severe complication: HFMD, acute hemorrhagic conjunctivitis, myocarditis, aseptic meningitis, severe neonatal sepsis-like disease, acute flaccid paralysis, and so on.17 The best known members of the genus Enterovirus are PV, EV7, CV, and echovirus. PV is spread via the fecal–oral route and rarely invades the central nervous system (CNS). However, in the rare instances when PV invades the CNS, the resulting damage to motor neurons is striking and often permanent. EV71 and CVA16 are common etiological agents of HFMD.18 As a neurotropic enterovirus, EV71 can violate the patient's nervous system and lead to severe symptoms and even to death.19,20 CV is classified into two types, CVA and CVB. CVAs are usually prevalent in summer and mainly cause herpetic angina, skin rash, meningitis, paralytic polio lesions, fetal intrauterine infection, and suffocation during infection in pregnancy. Some of CVAs, such as CVA16, CVA4, CVA5, CVA9, CVA10, are causative agents of HFMD as EV71. CVBs are the major pathogens of human viral myocarditis that can result in severe cardiac failure and dilated cardiomyopathy.21,22 Besides these, echoviruses 4 (E4), E16, and E30 are associated with type I diabetes.23 E4, E6, E11, E13, and E30 are related to acute meningitis/encephalitis,24,25 while E13 and E11 can cause non-polio acute flaccid paralysis.26
Accumulated evidence has demonstrated that there are intricate interactions between miRNAs and viruses. MiRNAs can be induced and regulated by viruses. For instance, miR-101 may serve as a potential biomarker in hepatocellular carcinoma (HCC) patients because miR-101 is down-regulated in Hepatitis B virus (HBV)-related HCC tissues.27 HBV X protein can inhibit miR-205 by hypermethylating of miR-205 promoter.28 Meanwhile, many evidence has showed that cellular miRNAs play a crucial role in regulating viral replication and pathogenesis in eukaryotes. For example, hsa-miR-29a down-regulates the expression of HIV Nef protein and interferes with HIV-I replication.29 MiR-198 can inhibit HIV-1 gene expression and replication in monocytes.30 Representatively, miR-122 positively regulates HCV infection by increasing HCV translation and genomic RNA stability through direct interactions with the viral RNA genome.31–33 At the same time, many researchers have demonstrated that miRNAs play an important role during EV infection. However, there are no summary about the role of miRNAs involved in enteroviral infection yet. Thus, this review attempts to provide recent advancements about the role and machinery of miRNAs in enteroviral infection, in order to increase our understanding of enteroviral pathogenesis.
The related investigations of miRNAs and EV71Due to the virulent pathogenicity of EV71, many researches have been carried out to explore the role of miRNAs during EV71 infection. It has been verified that circulating miRNAs play an essential role in pathogen–host interactions. Disruption of miRNA biogenesis by knocking down the DGCR8 gene, an essential cofactor for miRNAs biogenesis, resulted in the inability of EV71 to cause infection. The expression of many key genes related to the innate antiviral immune response in EV71 infected miRNAs depleted cells was significantly higher in comparison with control cells. It has been showed that EV71 can utilize host miRNAs to mediate host immune system enhancing infection.34 MiRNA can serve as a molecular marker for the detection of enteroviral infections. Cui and colleagues found that the expression level of 64 miRNAs changed over 2-fold in response to EV71 infection by deep sequencing assay, and 42 of these 64 miRNAs were up-regulated and others were down-regulated. Many of these miRNAs are associated with metabolic process and neurological processes, apoptosis, and immune response according to the result of gene ontology analysis.35 Subsequently, by using a miRNA array, this team reported that six miRNAs (miR-148a, miR-143, miR-324-3p, miR-628-3p, miR-140-5p, and miR-362-3p) in serum could discriminate patients with EV infections from healthy controls, and three miRNAs (miR-545, miR-324-3p, miR-143) could distinguish EV71 from CVA16 infections.36 Collectively, miRNAs are closely associated with enterovirual infection.
Some miRNAs can modulate the replication of EV71 by targeting the viral genome directly. For example, Zheng et al.37 found that miRNA-296-5p was elevated in EV71-infected cells by using a comprehensive miRNA profiling. They predicted by bioinformatic analysis that human miR-296-5p can inhibit the replication of EV71 by binding to the coding regions of VP3 and VP1 proteins. It is predicted that miR-23b can target the conservative sequence of EV71 3′ UTR by biological software. Then it is confirmed that miR-23b can inhibit the replication of EV71 by targeting EV71 3′ UTR by luciferase reporter vector and miR-23b mimics transfection.38 Yang et al.39 screened the miRNAs that may target EV71 gene sequence by using online analysis programs, then verified that miR373 and miR542-5p could suppress the replication of EV71 virus through binding to the 5′-UTR gene.
There are also some miRNAs that modulate viral replication indirectly by targeting the mediator. Interferon (IFN)-mediated pathway is a crucial part of the cellular response against viral infection. Type III IFNs, which include IFN-λ1, 2, and 3, mediate antiviral responses similar to Type I IFNs.40 There are some miRNAs that modulate viral replication involved IFN. MiRNA-548 can enhance replication of EV71 and vesicular stomatitis virus (VSV) by targeting the 3′UTR of IFN-λ1 and the levels of endogenous miRNA-548 are down-regulated during viral infection.41 EV71-induced miR-146a can suppress the expression of IRAK1 and TRAF6 which are two major elements in IFN production. Thus, EV71-induced miR-146a can enhance viral pathogenesis by suppressing IFN production.42 Retinoic acid-inducible gene I (RIG-I) is an intracellular RNA virus sensor that induces type I interferon-mediated host-protective innate immunity against viral infection. There is a novel positive regulator of RIG-I signaling pathway related to miRNA and EV71.43 EV71-induced miR-526a positively regulates type I IFN production and inhibits viral replication. The enhancement of this antiviral immune response then derived from suppressing cylindromatosis (CYLD) expression by miR-526a. Remarkably, virus-induced miR-526a upregulation and CYLD downregulation can be blocked by EV71 3C protein. Cells with overexpressed miR-526a enhance the resistance to EV71 infection. In addition to this, Chang et al. investigated an apoptosis-oriented approach by using mRNA-miRNA profiling and bioinformatic analysis. They predict and verified that miR-146a and miR-370 could impact two major apoptosis-associated signaling pathways by decreasing Son of sevenless homolog 1 (SOS1) and growth arrest and DNA damage-inducible protein 45β (GADD45β) expression, respectively. MiR-146a and miR-370 coordinate to trigger cell apoptosis in EV71 infection.44 EIF4E is the cap-dependent translation initiation factor to shut off host protein synthesis. MiR-141 can facilitate EV infection including EV71, PV3 and CVB3 by targeting 3′-UTR of eIF4E.45 EV71 down-regulated the expression of miR-27a significantly. Further study showed that miR-27a could inhibit EV71 replication by targeting epidermal growth factor receptor (EGFR).46
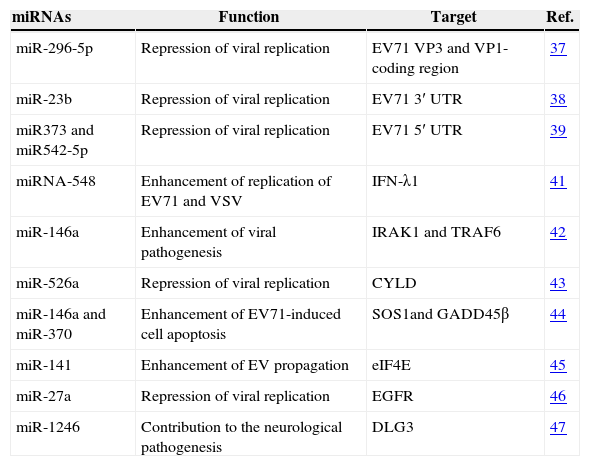
Moreover, to explain the neurological pathogenesis of EV71, Xu et al. did whole-genome joint mRNA and miRNA profile analysis from EV71-infected neuroblastoma cell SH-SY5Y. They found that miR-1246 was specifically up-regulated by EV71 in SH-SY5Y cells and could directly bound with the 3′ UTR of disk-large homolog 3 (DLG3) gene, which is associated with neurological disorders.47 This result may partly explain the neurological pathogenesis of EV71. We summarize the functions and the target genes of the miRNAs involved in the infection of EV71 in Table 1.
MiRNAs involved in EV71infection.
| miRNAs | Function | Target | Ref. |
|---|---|---|---|
| miR-296-5p | Repression of viral replication | EV71 VP3 and VP1-coding region | 37 |
| miR-23b | Repression of viral replication | EV71 3′ UTR | 38 |
| miR373 and miR542-5p | Repression of viral replication | EV71 5′ UTR | 39 |
| miRNA-548 | Enhancement of replication of EV71 and VSV | IFN-λ1 | 41 |
| miR-146a | Enhancement of viral pathogenesis | IRAK1 and TRAF6 | 42 |
| miR-526a | Repression of viral replication | CYLD | 43 |
| miR-146a and miR-370 | Enhancement of EV71-induced cell apoptosis | SOS1and GADD45β | 44 |
| miR-141 | Enhancement of EV propagation | eIF4E | 45 |
| miR-27a | Repression of viral replication | EGFR | 46 |
| miR-1246 | Contribution to the neurological pathogenesis | DLG3 | 47 |
CV including CVA and CVB mainly leads to herpangina and human heart disease, especially viral myocarditis (VMC). Many researches have focused on miRNAs related to CVB3 infection because CVB3 is a common cause of myocarditis and dilated cardiomyopathy. To evaluate the usefulness of miRNAs in the pathogenesis of CVB3-induced VMC, Zhang et al. analyzed the paired expression profiles of miRNAs and mRNAs on heart tissues from CVB3-infected mice. They identified five distinctly expressed miRNAs (miR-146a, miR-21, miR-374, miR-29a* and miR-23a), which are involved in regulating several important innate immune and antiviral pathways. This study showed that miRNAs can regulate the pathogenesis of CVB3-induced VMC.48
There are two miRNAs which can directly regulate the replication of CVB3 by targeting the viral sequence. By using RNAhybrid 2.2 and miRanda 3.2a software, Linlin Wang et al. showed that there is a putative miR-342-5p target (nt 4989–nt 5015) in the 2C-coding region of CVB3. The experiment in vitro and a moderate miR-342-5p abundance in CVB infected-Balb/c mice validated that miR-342-5p can inhibit CVB3 replication by targeting its 2C-coding region.49 Soon after, this group reported that miR-10a* can significantly augment the biosynthesis and replication of CVB3 151 by targeting its nt6918–nt6941 sequence of the 3D-coding region. This article shows that 152 miRNAs could up-regulate the expression of the target genes.50
There are also many miRNAs which can indirectly regulate the replication of CVB3 through mediator. Zinc finger protein (ZFP)-148, as a transcription factor, can regulate expression of many cell cycle regulatory genes and act as antiviral agents for many viruses. CVB3 infection up-regulated miR-203 through the activation of PKC/AP-1 cascade. Then, miR-203 up-regulated CVB3 replication through targeting ZFP-148.51 MiR-155 is reduced by infiltrating inflammatory macrophages and T lymphocytes of both human and mice.52–54 MiRNA-155, -146b, and -21 were consistently and significantly upregulated during VMC both in mice and human. Inhibition of miR-155 by LNA-Anti-miR in vivo caused cardiac injury and dysfunction, decreased T lymphocyte activation during acute myocarditis because miR-155 can target PU.1 (an inhibitor of dendritic cell antigen presentation to T cells). It is showed miR-155 is an adverse mediator of cardiac immune activation after CVB3-induced VMC. In vivo inhibition of miRNA-155 after CVB3 infection attenuates myocardial inflammation and necrosis, so miRNA-155 can serve as a novel therapeutic target for VMC.53 CVB3-induced miR-126 regulated CVB3 replication through two signal pathways, cross-talk of ERK1/2 and Wnt/β-catenin pathways. By transfecting miRNA mimics and inhibitors, it is suggested that miR-126 can regulate the ERK1/2 signaling pathways by targeting EVH1 domain containing 1 (SPRED1). Besides, MiR-126 can regulate Wnt/β-catenin pathways by targeting lipoprotein receptor-related protein 6 (LRP6) and Wnt-responsive Cdc42 homolog 1(WRCh1), which enhanced viral cytopathogenicity. In short, miR-126 promotes CVB3 replication through mediating cross-talk of ERK1/2 and Wnt/β-catenin signal pathways by aiming at three specific targets: SPRED1, LRP6, and WRCH1.55 In the mouse model of CVB3-infected VMC, miR-1 measured by real-time PCR was upregulated upon CVB3 infection and its target gene Connexin 43 (Cx43) measured by western blotting was significantly down-regulated. MiR-1 plays an important role in the pathophysiology of VMC and interferes with cardiac function by targeting Cx43, the main protein forming gap-junction channels in ventricular myocardium.56
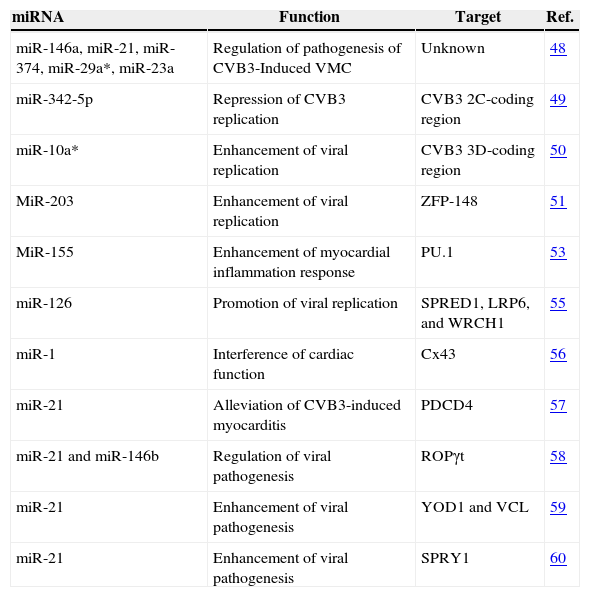
Interestingly, there are several studies about miRNA-21 involved in CVB3 infection. In CVB3 induced myocarditis, the expression of miR-21 was significantly decreased, while programmed cell death 4 (PDCD4) was increased. Further research showed that miR-21 alleviated CVB3-induced myocarditis and repressed myocardial apoptosis by targeting PDCD4.57 MiR-21 and miR-146b were upregulated in CVB3-infected VMC. Silencing of miR-21 and miR-146b can decrease the expression levels of Th17 and RORγt and result in less-severe damage. Further research showed that miR-21 and miR-146b can regulate pathogenesis of murine VMC by regulating TH-17 differentiation through targeting RORγt, TH-17 transcription factor.58 CVB3-upregulated miR-21 can trigger heart muscle cell damage by disrupting cardiomyocyte interactions. MiR-21 suppressed the levels of components in cell–cell interactions through targeting YOD1 and vinculin (VCL), which are two components related to the regulation of cell–cell connections and cardiac function. The miR-21 expression during CVB3 infection may contribute to the pathogenesis of VMC and the inhibition of miR-21 can reduce host injury.59 MiR-21 was significantly upregulated in cardiac myocytes from VMC and dilated cardiomyopathy (DCM) compared with control samples. By Using bioinformatics analysis, they predicted miR-21 could bind to 3′ UTR of homolog 1 (SPRY1). Further study in vitro revealed that the overexpression of miR-21 was associated with a decrease in SPRY1 protein expression. These results suggest that miR-21 may contribute to the pathogenesis of VMC to DCM by targeting SPRY1.60 At the same time, miRNA-21 was also related with other viruses by targeting a different gene. For example, miRNA-21 was correlated with herpes simplex virus (HSV)-induced Behçet's Disease in patients and in mouse model.61 Moreover, one of the HBV proteins, HBx can induce miRNA-21 expression in HBV-induced hepatocellular carcinoma. Then, HBV infection enhances cell proliferation via HBx-induced miRNA-21 by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN).62 We summarize the functions and the target genes of the miRNAs involved in the infection of CVB3 in Table 2.
MiRNAs involved in CVB3infection.
| miRNA | Function | Target | Ref. |
|---|---|---|---|
| miR-146a, miR-21, miR-374, miR-29a*, miR-23a | Regulation of pathogenesis of CVB3-Induced VMC | Unknown | 48 |
| miR-342-5p | Repression of CVB3 replication | CVB3 2C-coding region | 49 |
| miR-10a* | Enhancement of viral replication | CVB3 3D-coding region | 50 |
| MiR-203 | Enhancement of viral replication | ZFP-148 | 51 |
| MiR-155 | Enhancement of myocardial inflammation response | PU.1 | 53 |
| miR-126 | Promotion of viral replication | SPRED1, LRP6, and WRCH1 | 55 |
| miR-1 | Interference of cardiac function | Cx43 | 56 |
| miR-21 | Alleviation of CVB3-induced myocarditis | PDCD4 | 57 |
| miR-21 and miR-146b | Regulation of viral pathogenesis | ROPγt | 58 |
| miR-21 | Enhancement of viral pathogenesis | YOD1 and VCL | 59 |
| miR-21 | Enhancement of viral pathogenesis | SPRY1 | 60 |
| miRNA | Verified functions of the miRNA | Verified targets | Ref. |
|---|---|---|---|
| miR-342-5p | Repression of viral replication | CVB3 2C-coding region | 38 |
| miR-10a* | Enhancement of viral replication | CVB3 3D-coding region | 39 |
| MiR-203 | Enhancement of viral replication | ZFP-148 | 40 |
| MiR-155 | Enhancement of myocardial inflammation response | PU.1 | 42 |
| miR-126 | Promotion of viral replication | SPRED1, LRP6, and WRCH1 genes | 44 |
| miR-1 | Involved in viral myocarditis | Cx43 | 45 |
| miR-21 | Alleviation of CVB3-induced myocarditis | PDCD4 | 46 |
| miR-21 and miR-124b | Regulation of viral pathogenesis | ROPγt | 47 |
| miR-21 | Enhancement of viral pathogenesis | YOD1 and VCL | 48 |
| miR-21 | Enhancement of viral pathogenesis | SPRY1 | 49 |
There have been many recent reports about miRNAs related to EV71 and CVB3, including about miRNAs other EV, such as PV and HRV. The expression of miR-141 is upregulated in cells infected with PV3 and CVB3, besides EV71.45 MiR-141 can shut down host protein synthesis via mediating elF4E in PV3 and CVB3 infected cells, because eIF4E is the cap-dependent translation initiation factor for shutting off host protein synthesis.34 The replication of HRV is regulated by miRNAs as decrease of mature miRNAs reduced by DICER knock-down increases of HRV-1B replication in human bronchial epithelial cells. Specific miRNAs binding to viral RNA during HRV infection is demonstrated by co-immunoprecipitate test with argonaute 2 protein. Finally, out of this group of miRNAs, miR-128 and miR-155, under-expressed in the asthmatic epithelium, can affect HRV-1B replication.63 Innate immune responses can restrict viral replication and innate immune receptors, including toll-like receptor and retinoic acid inducible gene I (RIG-I)-like receptor (RLR), which activate IFN system. By using a microarray, several special miRNAs were found to be regulated by RLR signaling. One of these RLR-inducible miRNAs, miR-23b, targets very low density lipoprotein receptor (VLDLR) and LDLR-related protein 5 (LRP5). Overexpression of miR-23b and anti-miR-23b resulted in down-and up-regulated production of RV1B, respectively. It has been shown that RIG-I signaling results in inhibition of HRV1B infection because RIG-I-inducible miR-23 targets VLDLR, a receptor for viral entry into the cell.64
ConclusionsEmerging evidence has demonstrated that miRNAs play a key role in interaction network between the EV and host. EV infections can change the expression of miRNAs in the cell. Therefore, miRNAs affect enteroviral replication and pathogenesis. Some miRNAs can regulate the expression of EV gene directly; some miRNAs can regulate EV replication indirectly by targeting a mediator. When miRNAs directly act on enterovirus, miRNAs can combine with the 3′UTR, 5′UTR, VP1, VP3, 2C-coding region, 3D-coding region of EV to promote or suppress EV replication. When miRNAs indirectly interact with EV, miRNAs can play a regulatory role on virus through IFN, ZFP-148, or some other immune signaling pathways and so on. In conclusion, there are a variety of complex regulated relationship between miRNAs and EV, and miRNAs hold an important biological role in enteroviral infection.
Although many miRNAs are involved in the regulation of enteroviral infection, there is still incomplete understanding about this regulatory network. Based on the above review, miRNAs can bind to some regions of EV such as 3′UTR, 5′UTR, VP1, VP3, 2C-coding region, 3D-coding region. Are there some other EV targets of miRNAs? Although many studies have been reported about the process of interaction between miRNAs and EV71, CVB3, researches about miRNAs and other EV are still limited and more researches are needed. In addition, future studies are required to explore the precise mechanism by which miRNAs interfere in enteroviral infection in order to increase our understanding of the interactions between EV and miRNA. For example, EV71 can cause HFMD and lead to the development of severe neurological diseases. However, the involvement of miRNAs during the development of severe neurological diseases is still unclear.
Currently, more and more miRNAs have great potential usefulness to serve as biomarkers for the diagnosis of different diseases, such as cardiovascular disease,65 glioblastoma,66 and so on. Five miRNAs (miR-197, miR-629, miR-363, miR-132 and miR-122) could distinguish varicella patients from healthy controls and from patients with Bordetella pertussis, measles virus and EV.67 Importantly, miRNAs have a potential therapeutic role; SPC3649, a target of miR-122, is the first miRNAs-related drug to be tested in humans with hepatitis C.68 This discovery open up the possibility to develop miRNAs-related drugs to tackle enteroviral diseases. Future research is warranted to explore the regulatory network of the miRNAs and enterovirus, in order to explore miRNAs in the diagnosis and treatment of enteroviral infection.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by Science and Technology Support Program (Social Development) of Zhenjiang (Grant No. SH2012052), Jiangsu Province “333” Project.