Helicobacter pylori harbouring cag-pathogenicity island (cagPAI) which encodes type IV secretion system (T4SS) and cagA virulence gene are involved in inflammation of the gastric mucosa. We examined all the 27 cagPAI genes in 88 H. pylori isolates from patients of different ethnicities and examined the association of the intactness of cagPAI region with histopathological scores of the gastric mucosa.
Results96.6% (n=85) of H. pylori isolates were cagPAI-positive with 22.4% (19/85) having an intact cagPAI, whereas 77.6% (66/85) had a partial/rearranged cagPAI. The frequency of cag2 and cag14 were found to be significantly higher in H. pylori isolated from Malays, whereas cag4 was predominantly found in Chinese isolates. The cag24 was significantly found in higher proportions in Malay and Indian isolates than in Chinese isolates. The intactness of cagPAI region showed an association with histopathological scores of the gastric mucosa. Significant association was observed between H. pylori harbouring partial cagPAI with higher density of bacteria and neutrophil activity, whereas strains lacking cagPAI were associated with higher inflammatory score.
ConclusionsThe genotypes of H. pylori strains with various cagPAI rearrangement associated with patients’ ethnicities and histopathological scores might contribute to the pathogenesis of H. pylori infection in a multi-ethnic population.
Helicobacter pylori is a Gram-negative, microaerophilic, curved-shaped and flagellated bacterium frequently found in the stomach of humans.1 It is an important pathogen that causes gastrointestinal diseases such as chronic gastritis, peptic ulcer, gastric cancer and gastric mucosa-associated lymphoid tissue (MALT) lymphoma,2,3 and most infected patients appear asymptomatic. Although H. pylori is designated as type I carcinogen, other factors including environment (lifestyle and diet), host genetics and host immune responses contribute to the disease sequelae in infected patients.4–6
Cytotoxin-associated gene pathogenicity island (cagPAI) is one of the major virulence factors associated with disease outcome in infected hosts. It is approximately 40kb in size consisting of around 28 genes,7 encoding mainly CagA protein, type IV secretion system (T4SS) and other genes for induction of host’s interleukin-8 (IL-8).7,8 Studies show that intactness of cagPAI has a significant correlation with disease severity, whereas H. pylori strains with partial deletions within cagPAI region are significantly less pathogenic in nature.9,10 However, the rates of severe disease development vary between human populations, and differences in H. pylori genotypes may partially explain these differences.11,12
Integrity of cagPAI seems to have an important role in the progress of the gastroduodenal disorders, so that intact cagPAI could be seen in H. pylori strains from countries with higher rate of gastric cancer.13–15 This integrity also has important effect on eliciting inflammatory response in the gastric mucosa.16 Several studies have investigated the association of H. pylori cagPAI and gastroduodenal diseases.13,17 However, knowledge about the relationship between H. pylori cagPAI intactness and changes of the infected gastric tissue is sparse. More than 90% of H. pylori strains in Malaysia are cagPAI-positive18 and Malaysia being a multi-ethnic country, the interaction of H. pylori strains with different genotype combined with various host genetics may have an impact on associated disease outcomes.
There is a lack of comprehensive information with regarding intact versus rearranged cagPAI among H. pylori strains in the Malaysian population. Hence, in this study, we characterized the genes within cagPAI to determine the association of various cagPAI structure in H. pylori isolates with histopathological changes of the infected gastric mucosa. The outcome of this study provides valuable conclusions regarding association between presence of cagPAI genes and disease sequelae in strains from multi-ethnic population and associations with different histopathological states.
MethodsBacterial isolatesA total of 88 non-repetitive H. pylori clinical isolates were obtained from patients (47 females and 41 males) recruited in previous studies (research no. ETP-2013-042 and GUP-2011-307) between 2011–2015. Patients (15 Malays, 52 Chinese and 21 Indians) had a mean age of 54.68±16.87 years ranging from 17 to 83 years. In these projects, gastric biopsies were taken from the antrum and/or corpus of the patients’ stomach for H. pylori culture and used as one of the diagnotic methods for determination of the H. pylori infection status in patients. H. pylori isolates grew from the culture method during the studies were stored at −70°C in Brucella broth containing 15% glycerol. Histopathological examination was performed on gastric biopsies in both studies to detect the presence of H. pylori along with grading for gastritis. H. pylori were subcultured from frozen stock onto Columbia blood agar (Oxoid, Basingstoke, England) supplemented with 7% sheep blood and Dent’s supplement (Oxoid, Basingstoke, England) and incubated at 37°C for five to seven days under microaerophilic environment.
Histopathological examinationGastric biopsies fixed in 10% formalin and paraffin embedded section were cut and stained with hematoxylin-eosin and, when necessary, sections were also stained with Warthin-Starry for better visualization of H. pylori. All patients had gastritis graded according to Updated Sydney Classification31 except in two patients where the histopathological examination (HPE) results were not available. Severity of gastritis was graded from 0 to 3 (none, mild, moderate, and marked).
DNA extractionH. pylori colonies were scraped from the agar surface of Columbia blood agar plate and subjected to DNA extraction using FavorPrep™ Tissue Genomic DNA Extraction Mini kit according to the manufacturer’s instructions (Favorgen Biotech Corporation, Ping-Tung 908, Taiwan). DNA samples were diluted with ultrapure water to a concentration of 25ng/μl and stored at -20°C until further processing.
Determination of cagPAI genesThe presence or absence of cagPAI in H. pylori strains was determined by PCR using primers for detection of the 5′ and 3′ flanking region of the cagPAI as described by Olbermann et al.28 The amplifications were carried out in 25μL volume, each containing 12.5μL mastermix (Lucigen, USA), 10μL of each primers, 1μL (25ng) DNA and 10μL DNAse and RNAse free sterile distilled water. PCR amplification for detection of cagPAI region consisted of initial denaturation at 95°C for 3min, followed by 30 cycles of 95°C for 30s, 50°C for 60s, and 72°C for 45s, ending with final extension at 72°C for 5min. The amplifications were performed in a PCR thermal cycler T100 Series (Bio-Rad, USA). The products were run on 1.5% agarose gel and stained with FloroSafe DNA stain (1st BASE Pte. Ltd, Singapore) and visualized with gel documentation (AlphaImager, Biosciences, CA). The cagPAI-positive isolates (n=91) were then subjected to subsequent PCRs for identification of all cagPAI genes using primers as described previously.28,32 The absence of cag2 was confirmed with 690 or 1100 bp amplicon using empty-site PCR.24cag14 was detected using four sets of primer pair as described earlier.32 PCR amplification for cagPAI genes consisted of initial denaturation at 95°C for 3min, followed by 30 cycles of 95°C for 30s, annealing temperature for 60s (48°C for cag11, 48.8°C for cag3 and 55°C for cag1, cag2, cag4, cag5, cagα, cag6 to cag10, cag12 to cag26), and extension at 72°C for 45s. A final extension at 72°C for 5min was performed for each PCR run. Representative positive PCR products (n=28) were sent for sequencing and the nucleotide sequences were blasted against NCBI databases to confirm the gene identity.
Statistical analysisStatistical analysis was performed using SPSS software version 23 (SPSS Inc, Chicago, IL, USA). Differences between groups were evaluated using Chi-square (χ2) test with Yate’s continuity correction and Fisher’s exact test. Independent t-test was used to compare means between different groups of histopathological scores. Score was represented with mean±standard error of mean (SE). Differences were considered significant when p value was <0.05.
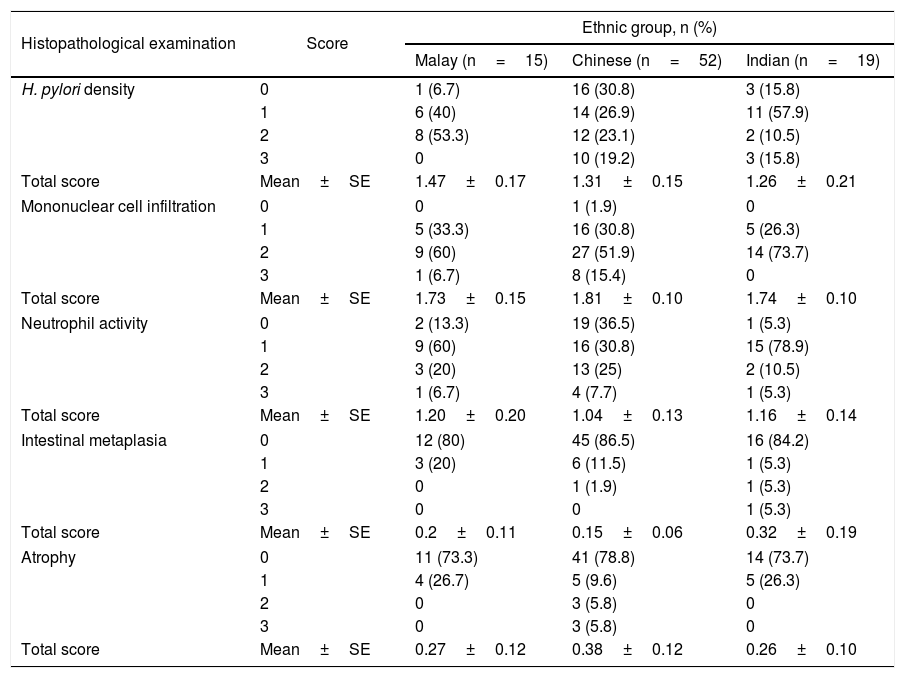
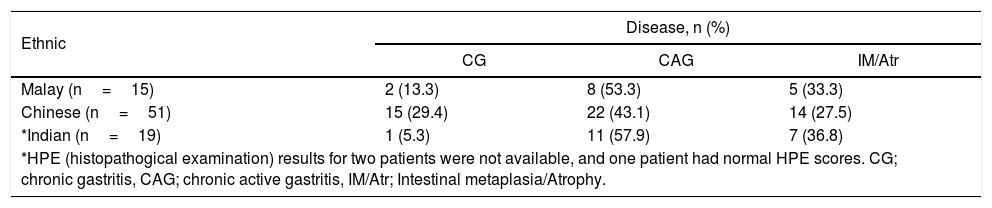
ResultsHistopathological characteristics of the gastric mucosa in the studied ethnic groupsHistopathological scores of the gastric mucosa among different ethnic groups showed that the Malays had higher mean scores for H. pylori density and neutrophil activity whereas the Chinese showed higher grade of inflammation (Table 1). Higher mean score for intestinal metaplasia was observed among the Indians, while atrophy of higher grade was observed in the Chinese. Patients of different ethnicities were grouped into different types of disease conditions based on the histopathological changes (Table 2), i.e. chronic gastritis (CG) (n=18), chronic active gastritis (CAG) (n=41) and intestinal metaplasia/atrophy (IM/Atr) (n=26). There was a significant difference in the proportion of CG and CAG between Chinese and non-Chinese patients. CG was more diagnosed in Chinese patients compared to non-Chinese (p=0.03), whereas CAG and IM/Atr were more observed in non-Chinese than in Chinese (p=0.042) patients.
Histopathological scores of the gastric mucosa in different ethnic groups.
| Histopathological examination | Score | Ethnic group, n (%) | ||
|---|---|---|---|---|
| Malay (n=15) | Chinese (n=52) | Indian (n=19) | ||
| H. pylori density | 0 | 1 (6.7) | 16 (30.8) | 3 (15.8) |
| 1 | 6 (40) | 14 (26.9) | 11 (57.9) | |
| 2 | 8 (53.3) | 12 (23.1) | 2 (10.5) | |
| 3 | 0 | 10 (19.2) | 3 (15.8) | |
| Total score | Mean±SE | 1.47±0.17 | 1.31±0.15 | 1.26±0.21 |
| Mononuclear cell infiltration | 0 | 0 | 1 (1.9) | 0 |
| 1 | 5 (33.3) | 16 (30.8) | 5 (26.3) | |
| 2 | 9 (60) | 27 (51.9) | 14 (73.7) | |
| 3 | 1 (6.7) | 8 (15.4) | 0 | |
| Total score | Mean±SE | 1.73±0.15 | 1.81±0.10 | 1.74±0.10 |
| Neutrophil activity | 0 | 2 (13.3) | 19 (36.5) | 1 (5.3) |
| 1 | 9 (60) | 16 (30.8) | 15 (78.9) | |
| 2 | 3 (20) | 13 (25) | 2 (10.5) | |
| 3 | 1 (6.7) | 4 (7.7) | 1 (5.3) | |
| Total score | Mean±SE | 1.20±0.20 | 1.04±0.13 | 1.16±0.14 |
| Intestinal metaplasia | 0 | 12 (80) | 45 (86.5) | 16 (84.2) |
| 1 | 3 (20) | 6 (11.5) | 1 (5.3) | |
| 2 | 0 | 1 (1.9) | 1 (5.3) | |
| 3 | 0 | 0 | 1 (5.3) | |
| Total score | Mean±SE | 0.2±0.11 | 0.15±0.06 | 0.32±0.19 |
| Atrophy | 0 | 11 (73.3) | 41 (78.8) | 14 (73.7) |
| 1 | 4 (26.7) | 5 (9.6) | 5 (26.3) | |
| 2 | 0 | 3 (5.8) | 0 | |
| 3 | 0 | 3 (5.8) | 0 | |
| Total score | Mean±SE | 0.27±0.12 | 0.38±0.12 | 0.26±0.10 |
Disease groups in different ethnicities.
| Ethnic | Disease, n (%) | ||
|---|---|---|---|
| CG | CAG | IM/Atr | |
| Malay (n=15) | 2 (13.3) | 8 (53.3) | 5 (33.3) |
| Chinese (n=51) | 15 (29.4) | 22 (43.1) | 14 (27.5) |
| *Indian (n=19) | 1 (5.3) | 11 (57.9) | 7 (36.8) |
| *HPE (histopathogical examination) results for two patients were not available, and one patient had normal HPE scores. CG; chronic gastritis, CAG; chronic active gastritis, IM/Atr; Intestinal metaplasia/Atrophy. | |||
| Statistical analysis: | ||
|---|---|---|
| Ethnic | Disease, n (%) | |
| CG | CAG | |
| Non-Chinese (n=22) | 3 (13.3) | 19 (86.4) |
| Chinese (n=37) | 15 (40.5) | 22 (59.5) |
| *Non-Chinese (Malay and Indian), χ2=4.710, df=1, p= 0.03. | ||
| Ethnic | Disease, n (%) | |
|---|---|---|
| CG | IM/Atr | |
| Non-Chinese (n=15) | 3 (20) | 12 (80) |
| Chinese (n=29) | 15 (51.7) | 14 (45.3) |
| χ2=4.116, df=1, p= 0.042. | ||
A total of 96.6% (n=85) of the isolates were cagPAI-positive. Five genes in the cagPAI region (cag1, cag5, cag6, cag8 and cag21) were detected in all isolates whereas cag2 was detected in 34.1% (n=29) and cag14 in 51.7% (n=44) of the isolates (Table S1). Detection of other genes ranged from 69.4 to 98.8%.
Six genes (cag1, cag5, cag6, cag8, cag21 and cag26) in the cagPAI region were detected in all Indian isolates, whereas 12 and 19 genes were detected in all Chinese and Malay isolates, respectively (Table S1). The twelve genes detected in Chinese isolates were cag1, cag3, cag5, cag6, cag8, cag9, cag12, cag13, cag15, cag20-22. The 19 genes detected in Malay isolates were cag1, cag3, cag5-9, cag11-13, cag15-19, cag21-23 and cag26. A significant difference in detection of cag2, cag4, cag14 and cag24 were observed among H. pylori from patients with different ethnicities. Detection of cag2 was significantly higher in isolates from Malays (86.7%), followed by Indians (57.9%) and was least in Chinese isolates (9.8%) (χ2=36.62, df=2, p<0.0001). The presence of cag4 was more frequent in isolates from Chinese (80.4%) compared to the Malays (46.7%) and Indians (63.2%) (χ2=7.001, df=2, p=0.03). Significant difference was observed in the detection of cag14 in Malay isolates (93.3%) compared to Chinese (39.2%) and Indian (52.6%) isolates (χ2=13.603, df=2, p=0.001). Also, the frequency of cag24 was significantly higher in isolates from Malays (93.3%) and Indians (89.5%) compared to isolates from Chinese patients (54.9%) (χ2=12.701, df=2, p=0.002).
We did further analyses to look for the distribution of individuals cagPAI genes in different disease conditions. All cagPAI genes showed a similar distribution in CG, CAG and IM/Atr (data not shown) except for the cag2. cag2 was detected in 11.8% (2/17) of CG, 35% (14/40) of CAG and 44% (11/25) of IM/Atr. However, no significant difference was observed for the detection of H. pylori carrying cag2 in different groups of diseases (p=0.086).
Analysis of cagPAI intactness in H. pylori isolatesThe cagPAI was defined as intact if all gene sets of the cagPAI were present including strains lacking only the cag2 (HP0521). A previous systematic mutagenesis study showed that the HP0521 gene was not involved in the process of CagA translocation and IL-8 induction.7 In addition, NCBI database defined the HP0521 as a pseudogene (NCBI-Gene ID: 900040) (DBGET/LinkBD: an integrated database retrieval system, last accessed Oct 8, 2018). Partial cagPAI was defined when an isolate lacked one or more cagPAI genes other than cag2 (HP0521), while negative/deleted cagPAI was defined if none of the genes was present and a product of approximately 650bp with primers from the flanking regions was obtained. Among the 85 cagPAI-positive H. pylori strains, 22.4% (n=19) had intact cagPAI and 77.6% (n=66) exhibited partial (rearranged) cagPAI. Strains harbouring intact or partial cagPAI were not associated with patients’ ethnicities (p>0.05).
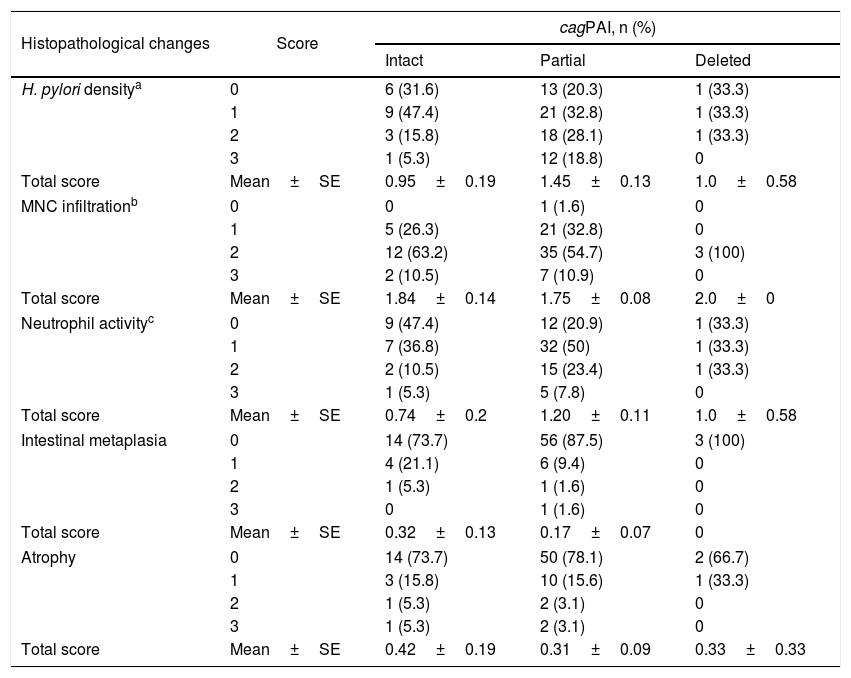
Association between cagPAI intactness and histopathological scores of the gastric mucosa are shown in Table 3. The presence of partial cagPAI was significantly related to higher total score of H. pylori density (p=0.037) and neutrophil activity (p=0.038) compared to intact cagPAI. H. pylori harbouring deleted cagPAI was significantly correlated with higher inflammatory score (mononuclear infiltration) compared to H. pylori with partial cagPAI (p=0.004). The distribution of H. pylori with intact cagPAI was more detected in the gastric mucosa with IM/Atr, whereas partial cagPAI H. pylori was more detected in CAG, however the difference was not significant. Moderate and severe scores of H. pylori density, mononuclear infiltration, neutrophil activity, and atrophy were observed more in gastric mucosa from Chinese patients infected with H. pylori strains harbouring cagPAI than other ethnicities (Malays and Indians) (Table S2).
Association of H. pylori cagPAI intactness with histopathological changes of gastric mucosa.
| Histopathological changes | Score | cagPAI, n (%) | ||
|---|---|---|---|---|
| Intact | Partial | Deleted | ||
| H. pylori densitya | 0 | 6 (31.6) | 13 (20.3) | 1 (33.3) |
| 1 | 9 (47.4) | 21 (32.8) | 1 (33.3) | |
| 2 | 3 (15.8) | 18 (28.1) | 1 (33.3) | |
| 3 | 1 (5.3) | 12 (18.8) | 0 | |
| Total score | Mean±SE | 0.95±0.19 | 1.45±0.13 | 1.0±0.58 |
| MNC infiltrationb | 0 | 0 | 1 (1.6) | 0 |
| 1 | 5 (26.3) | 21 (32.8) | 0 | |
| 2 | 12 (63.2) | 35 (54.7) | 3 (100) | |
| 3 | 2 (10.5) | 7 (10.9) | 0 | |
| Total score | Mean±SE | 1.84±0.14 | 1.75±0.08 | 2.0±0 |
| Neutrophil activityc | 0 | 9 (47.4) | 12 (20.9) | 1 (33.3) |
| 1 | 7 (36.8) | 32 (50) | 1 (33.3) | |
| 2 | 2 (10.5) | 15 (23.4) | 1 (33.3) | |
| 3 | 1 (5.3) | 5 (7.8) | 0 | |
| Total score | Mean±SE | 0.74±0.2 | 1.20±0.11 | 1.0±0.58 |
| Intestinal metaplasia | 0 | 14 (73.7) | 56 (87.5) | 3 (100) |
| 1 | 4 (21.1) | 6 (9.4) | 0 | |
| 2 | 1 (5.3) | 1 (1.6) | 0 | |
| 3 | 0 | 1 (1.6) | 0 | |
| Total score | Mean±SE | 0.32±0.13 | 0.17±0.07 | 0 |
| Atrophy | 0 | 14 (73.7) | 50 (78.1) | 2 (66.7) |
| 1 | 3 (15.8) | 10 (15.6) | 1 (33.3) | |
| 2 | 1 (5.3) | 2 (3.1) | 0 | |
| 3 | 1 (5.3) | 2 (3.1) | 0 | |
| Total score | Mean±SE | 0.42±0.19 | 0.31±0.09 | 0.33±0.33 |
Statistical analysis (Independent t-test):
aPartial vs Intact; t=2.173, p= 0.037, 95% CI (0.033−0.978).
Partial vs Deleted; p= 0.46.
Deleted vs Intact; p= 0.92.
bDeleted vs Partial; t=3.000, p= 0.004, 95% CI (0.083–0.417).
Deleted vs Intact; p= 0.661.
Intact vs Partial; p= 0.591.
cPartial vs Intact; t=2.108, p= 0.038, 95% CI (0.026–0.906).
Deleted vs Intact; p= 0.701.
Deleted vs Partial; p= 0.760.
Major differences in the prevalence of H. pylori infection and disease-related severity were observed among patients from multiracial ethnicities.19,20 Bacterial virulence factor is one of the contributing factors for the development of severe H. pylori-related diseases. The diversity of cagPAI organization in the H. pylori genome may have a modifying effect on the pathogenic potential of the infecting strain.21
In this study, we comprehensively determined the presence of cagPAI region in 88 H. pylori isolates from Malaysian population which were isolated from patients of different ethnic groups. The results showed that more than 95% of H. pylori strains were cagPAI-positive where 22.4% of the isolates carried all cagPAI genes and 77.6% exhibited partial or rearrangement in the cagPAI genes. Our previous study had showed that 3.2% of the isolates had all the cagPAI genes.18 Low percentage of H. pylori isolates harbouring intact cagPAI genes was observed in our previous study because only a subset of the cagPAI genes (cag67, cag10, cag13, cagT, cagM and cagE) was analysed as these genes were shown to have linkage with certain cagPAI genes and severe disease outcome as described in earlier studies.22,23 In contrast, high frequency of intact cagPAI and low frequency of partial cagPAI in H. pylori strains isolated from similar ethnic populations was reported by Schmidt et al.24 In their study, cagE, cagL, cagT and HP521 were examined to assess the intactness of cagPAI region. Discordance in the frequency of cagPAI intactness in many reports was due to the different cagPAI genes being examined.13,25,26 Thus, results of the present study indicate that deletions can occur in all parts of the cagPAI and screening the entire genes in the cagPAI is essential for an accurate determination of the organization of the cagPAI region. For comparison with our results, we reviewed only studies that screened all the cagPAI genes. A previous study observed complete cagPAI present in 82.6% of the strains, while a partially deleted cagPAI in 9.6% of the strains and 7.7% lacked the entire cagPAI in Indian population.10 In Swedish population, 76% of the strains carried an intact cagPAI, 15% had partially deleted cagPAI and 9% of the strains lacked the cagPAI.9 A study by Azuma et al.27 showed that the complete cagPAI was identified in all 11 Japanese isolates. Variation in the cagPAI positivity in different population of H. pylori isolates might be related to different geographical origin of H. pylori subpopulations. Presence of the cagPAI region is almost universal in H. pylori hpEastAsia and hpAfrica1 populations, intermediate presence in hpEurope, and complete absence in hpAfrica2.28 Malaysian isolates showed a mixed subpopulation of hpEastAsia, hpAsia2, and hpEurope as indicated by multiracial communities living in the country.29,30
Analysis of the entire 27 cagPAI genes in the present study revealed that cag1, cag5, cag6, cag8 and cag21 were present in all isolates, which might represent core genes of the cagPAI region. However, function of the cag1, cag6 and cag21 are still unknown.28cag5 (HP0524, cagβ) and cag8 (HP0528, cagX) is a component of T4SS (VirB9 and VirD4).28 One strain lacked cagA gene but had other cagPAI genes indicating that cagA-positive isolates do not necessarily harbour intact cagPAI region. Indian isolates showed more rearrangement in the cagPAI region compared to Malay and Chinese isolates. Studies show that the subpopulations of H. pylori Indian isolates in Malaysia consist of mixed populations i.e., hpEurope, hpAsia2, and hpEAsia reflecting on the diversity of cagPAI genes rearrangement in the Indian isolates.29,30
The presence of specific genes in H. pylori isolates associated with different ethnicities (cag4 in the Chinese isolates and cag2, cag14 and cag24 in the non-Chinese isolates) might represent strains associated disease outcomes. The cag4 (VirB1) is a component of T4SS, whereas the function is still unknown for cag2, cag14 and cag24.28 Although the difference was not statistically significant, high frequency of cag2 was detected in gastric mucosa with CAG and IM/Atr and reflects the presence of this gene in non-Chinese isolates. These observations require further investigation to decipher the role of these genes.
We found an association of cagPAI intactness with histopathological scores of the gastric mucosa. H. pylori harbouring partial cagPAI were associated with higher density of H. pylori and neutrophil activity, whereas H. pylori with deleted cagPAI caused increased in inflammatory score. The presence of neutrophil activity in the gastric mucosa was associated with CAG. In addition, in our study partial cagPAI H. pylori strains was more often detected in patients with CAG. As strains with deleted cagPAI only cause inflammation of the gastric mucosa, the presence of cagPAI proteins encoded by H. pylori strains is needed to cause more severe disease such as active gastritis and intestinal metaplasia. However, no specific gene causing severe conditions could be identified. A group of genes that encodes T4SS and induction of IL-8 secretion has been shown to be involved in disease development process.7,24
ConclusionsThis study shows the diversity in the arrangement of cagPAI in H. pylori isolates obtained from different ethnic groups in Malaysia. Comprehensive screening of the entire cagPAI genes provided a more accurate overview of the H. pylori cagPAI genotype allowing for better identification of virulence traits of H. pylori in a multiracial population. This has implications for assessing different treatment options for treating H. pylori infections and can affect various disease outcomes. H. pylori strains harbouring partial/rearrangement of the cagPAI genes were associated with increased colonization and recruitment of neutrophils at the site of infection and further contribute to various disease outcomes caused by different H. pylori genotypes. Further studies in this area not only provide more information about the disease prevalence in various ethnic groups in Malaysia but also inform healthcare practitioners if a person from an ethnic group may be at risk of developing severe disease leading to the development of a customized treatment plan. The data obtained can be used to monitor national trends and develop further appropriate intervention strategies.
FundingThe research was funded by a grant from Ministry of Higher Education of Malaysia (grant no. FRGS/2/2014/SKK04/UKM/02/01). We also thank to Ministry of Higher Education of Malaysia for providing a studentship to SAR under the MyBrain15 program.
Availability of data and materialsData will be shared upon request to the corresponding author alfizah@ppukm.ukm.edu.my.
Authors’ contributionSAR performed all experiments and data analysis. HMN and NMZ participated in the study design and data analysis. AH involved in the design of the study, data analysis and manuscript writing. BSL participated in data analysis and manuscript writing. All authors read and approved the final manuscript.
Ethics approval and consent to participateThe research protocol was approved by the Medical Research Ethic Committee of the University (UKM1.5.3.5/244/JEP-2016-095). The present study used H. pylori stock cultures where the informed consent was not applicable. However, these isolates were obtained from patients in previous studies (research no. ETP-2013-042 and GUP-2011-307) where informed consent was obtained from all individuals included in the studies.
Conflicts of interestThe authors declare no conflicts of interest.
We would like to thank to the Universiti Kebangsaan Malaysia for providing both the permission and the facilities to conduct and publish this research and to the technical staffs of Dept. of Medical Microbiology & Immunology, Faculty of Medicine, Universiti Kebangsaan Malaysia for their technical help.