Bacterial tonsillitis is an upper respiratory tract infection that occurs primarily in children and adolescents. Staphylococcus aureus is one of the most frequent pathogens in the etiology of tonsillitis and its relevance is due to its antimicrobial resistance and persistence in the internal tissues of the tonsils. Tonsillectomy is indicated in cases of recurrent tonsillitis after several failures of antibiotic therapy.
Material and methodsIn this study we evaluated 123 surgically removed tonsils from patients who had history of recurrent tonsillitis. The tonsils were submitted to microbiological analysis for detection of S. aureus. The isolates were identified by PCR for femA gene. Antimicrobial susceptibility of the isolates was determined by disk diffusion tests. All isolates were submitted to PCR to detect mecA and Panton-Valentine leucocidin (PVL) genes. The genetic similarity among all isolates was determined by pulsed field gel electrophoresis.
ResultsSixty-one S. aureus isolates were obtained from 50 patients (40.7%) with mean age of 11.7 years. The isolates showed high level resistance to penicillin (83.6%), 9.8% had inducible MLSb phenotype, and 18.0% were considered multidrug resistant (MDR). mecA gene was detected in two isolates and the gene coding for PVL was identified in one isolate. The genetic similarity analysis showed high diversity among the isolates. More than one genetically different isolate was identified from the same patient, and identical isolates were obtained from different patients.
ConclusionsMDR isolates colonizing tonsils even without infection, demonstrate persistence of the bacterium and possibility of antimicrobial resistance dissemination and recurrence of infection. A specific clone in patients colonized by S. aureus was not demonstrated.
Tonsillitis is an infection of the palatine tonsils that occurs mostly in children and young adults, and recurrent tonsillitis is among the most common childhood diseases.1,2 Much has been written about the etiology of recurrent tonsillitis, but it remains a controversial topic. While a single microbial species may cause acute tonsillitis, it has been suggested that recurrent tonsillitis is a consequence of a polymicrobial infection.2 Microorganisms other than Group A Beta Hemolytic Streptococcus (GABHS) may be the cause of chronic tonsillitis.1
Bacterial biofilms are recognized as the main factor involved in the chronicity of infections and resistance to antibiotic treatment. Therefore, these infections have a considerably negative impact on patients’ quality of life and a significant burden on public health. Bacterial biofilm may play a role in various recurrent/chronic upper respiratory tract infections, including chronic tonsillar disease. Biofilm has been found in the tonsillar tissue of children with chronic infections.3
Especially in recent years, the presence of the beta-lactamase producing bacteria such as Staphylococcus aureus and Haemophilus influenzae in tonsils microbiota can promote penicillin resistance. Several researchers have claimed that failure of antibiotic therapy may be due to the underestimation of resistant microorganisms,1 which could also be explained by low concentration of antibiotics in the tonsillar tissue, potentially combined with the presence of resident bacteria producing protective enzymes, or specific antibiotic resistance patterns of the involved pathogenic bacteria.4
The presence of the bacterium in the internal tissue of the tonsil may be responsible for its persistence in this site. Tonsillar surface commonly presents bacteria belonging to the normal oral microbiota, and internal tissue contains pathogenic microorganisms. S. aureus have been detected in both external and internal tissues of the tonsils.4
In this study, we investigated the prevalence of S. aureus from tonsils that were removed due to recurrent tonsillitis as well as the antimicrobial susceptibility profile of the isolates. In addition, virulence genes were investigated, and the genetic similarity among the isolates was also determined.
Material and methodsStudy setting and ethics statementThis cross-sectional study was carried out at a tertiary hospital, in the Department of Surgical Clinic of a Teaching Hospital in Goiania, a municipality in midwest Brazil. The study protocol was approved by the local Ethics Committee (Protocol CEP/HC/UFG number 071/2011), and written informed consent was obtained from the children's parents or legal guardians.
Study subjectsFor two years, outpatients aged 0.8–48 years old who were referred to the Department of Surgical Clinic to perform tonsillectomy were recruited. The patients had a history of tonsillar hyperplasia and/or recurrent tonsillitis after repeated failures of antimicrobial therapy.
Following tonsillectomy, the tonsils were placed in a sterile container and promptly taken to the Medical Bacteriology Laboratory of the Federal University of Goias, where they were processed.
Samples processingAll of the samples were homogenized in Buffered Peptone Water 0.1% and inoculated in Mannitol Salt agar and Sheep's Blood agar (5%) for bacterial isolation. The isolates were identified according to standard methods.5 Isolates presenting coccus morphology, Gram positive stain, beta hemolysis, and producing catalase, coagulase, and DNAse were submitted to PCR for identification of S. aureus by detection of femA gene.
DNA extrationGenomic DNA was extracted from cultures grown on Tryptic Soy agar plates and was then used as a template for amplification according to Aires de Souza et al.6
PCR for detection of femA geneThe isolates were screened by PCR for the presence of femA gene, which is specific for S. aureus, according to Mehrotra et al. (2000).7 The primers used for gene amplification were femA-F: 5′ AAAAAAGCACATAACAAGCG 3′ and femA-R: 5′ GATAAAGAAGAAACCAGCAG 3′, to obtain a 132bp amplicon.
Determination of antimicrobial susceptibility profileThe antimicrobial susceptibility profile of the S. aureus isolates to penicillin, cefoxitin, erythromycin, clindamycin, quinupristin–dalfopristin, linezolid, trimethoprim-sulfamethoxazole, amoxicillin-clavulanate, ciprofloxacin, ceftriaxone, tetracycline, gentamicin, and rifampin was determined by disk diffusion test according to the Clinical Laboratory Standards Institute (CLSI) guidelines.8 The S. aureus strain ATCC 25923 was used as control. To determine the inducible or constitutive resistance profile to macrolides, lincosamides, and streptogramins B (MLSb), the D-zone test was performed according to CLSI.8
All methicillin-resistant S. aureus (MRSA) and the isolates that showed resistance to three or more antimicrobial classes were considered multidrug-resistant (MDR).9
PCR for detection of mecA geneThe detection of mecA gene was considered as MRSA marker. The primers used to amplify the mecA gene (310bp) were P1: (5′-TCCAGATTACAACTTCACCAGG-3′ and P2: 5′-CCACTTCATATCTTGTAACG-3′) as previously described.10
PCR for detection of Panton-Valentine LeukocidinThe primers were designed to obtain amplification of lukS-PV and lukF-PV genes according to the published sequences (GenBank accession numbers X72700 and AB006796).11
The amplified PCR products were electrophoresed in 1.5% agarose gels in TBE (Tris Borate EDTA) buffer at 120V for 1h, stained with ethidium bromide, and photographed under UV Molecular Imager® GelDoc™ XR system (Bio-Rad Laboratories, Hercules, CA, USA).
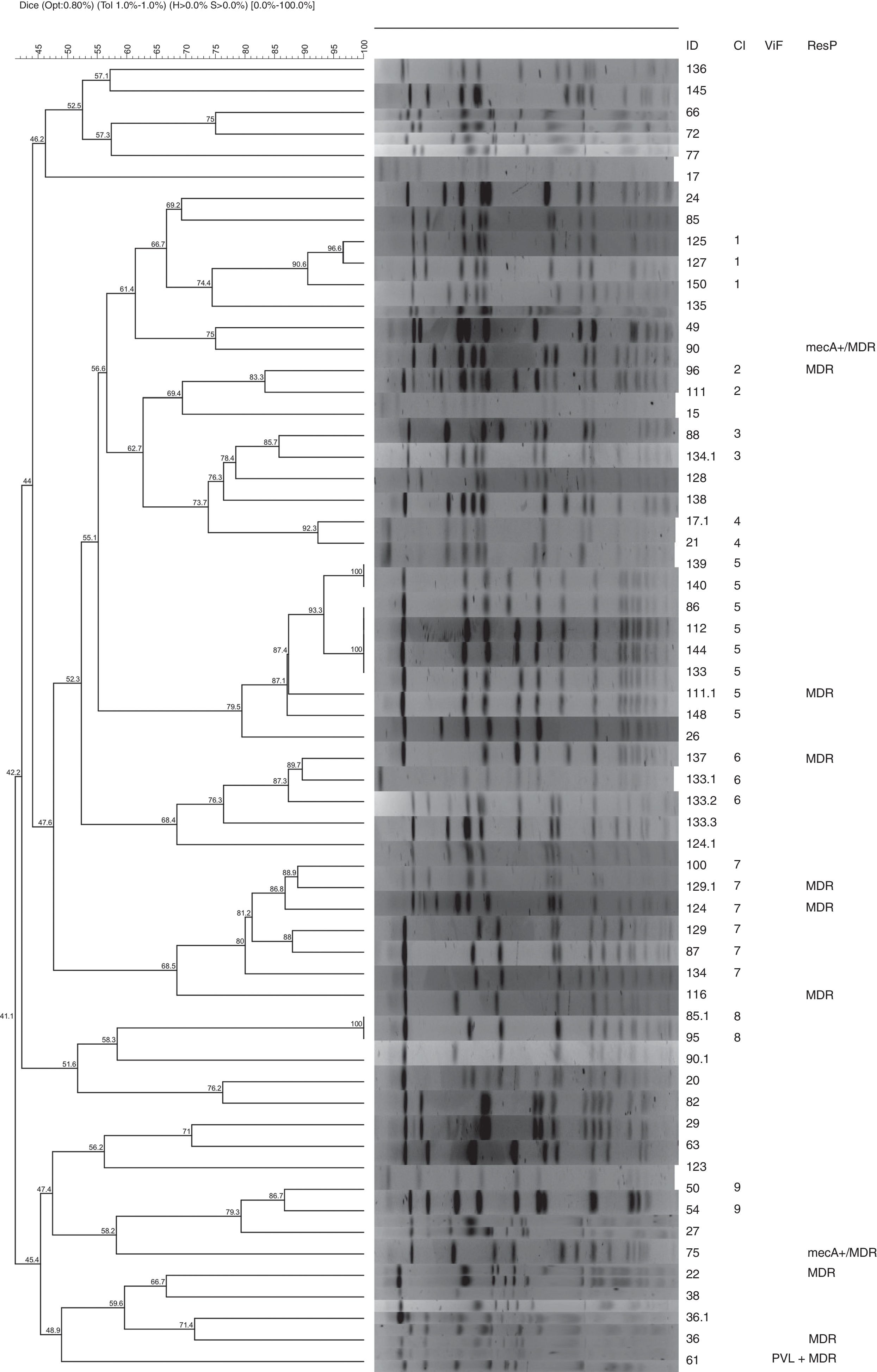
Pulsed field gel electrophoresis – PFGEPulsed-field gel electrophoresis (PFGE) of SmaI-digested macro fragments of the respective chromosomal DNA was used to fingerprint the isolates according to methods described previously,12 using 20U SmaI restriction enzyme. Electrophoretic run was performed in 0.5X TBE buffer (90mM Tris, 90mM Boric acid, and 2mM EDTA) under the following conditions: 6V/cm for 22h, pulse times from 5 to 35s at 14°C, using CHEF DRII System (Bio-Rad Laboratories, Hercules, CA, USA). The chromosomal DNA restriction patterns were analyzed with Bionumerics software (version 5.0; AppliedMaths, Ghent, Belgium) using the UPGMA algorithm with Dice correlation coefficients (0.8% optimization and 1.0% tolerance setting). Identical PFGE profiles (100% of similarity) were defined as a pulsotype (PT), and a cut-off of 80% was applied to cluster the strains.
Statistical analysisThe descriptive statistics were analyzed with Statistical Package for the Social Sciences (SPSS) version 17.0 (SPSS, Chicago, IL, USA) and Epi Info software version 2000 (CDC, Atlanta, GA, USA). Categorical variables were compared by chi-square or Fisher's exact test, when appropriate. Differences in means were assessed by the Student T-test. p≤0.05 was deemed statistically significant.
ResultsTonsils from 123 patients, aged 0.8–48 years, were analyzed over two years. The mean age of the patients was 11.3 years, of whom 53.7% (n=66) were male and 46.3% (n=57) female.
Of the 123 tonsils analyzed, 61 isolates (49.6%) were identified as S. aureus by femA gene detection. It should be noted that at the time of the tonsillectomy, the patients had no acute inflammatory process.
S. aureus was isolated in 50 out of 123 (40.7%) patients aged 0.8–36 years (mean age=11.7 years), in whom 76.0% (n=38) had recurrent pharyngotonsillitis and 88.0% (n=44) presented tonsillar hypertrophy, with degrees of obstruction varying between III and IV. In 12.0% (6/50) of the patients, S. aureus was the only agent found, and in 18.0% (9/50) two or more S. aureus isolates with different genotypic profiles were identified. Of these 50 patients, 78.0% reported antimicrobial use before tonsillectomy. The drug of choice to treat the pharyngotonsillitis was amoxicillin, which was used by 44.0% (22/50) of the patients.
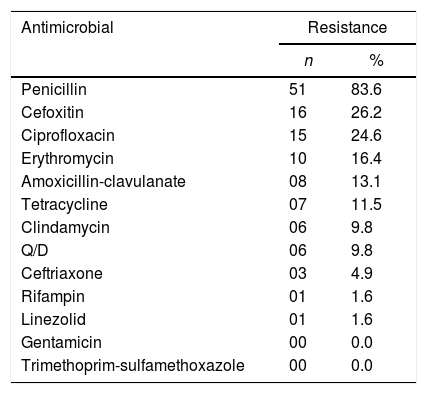
According to the antibiogram, the S. aureus isolates presented greater resistance to penicillin (83.6%) followed by cefoxitin (26.2%), ciprofloxacin (24.6%) and amoxicillin-clavulanate (13.1%). Six isolates (9.8%) presented inducible MLSb phenotype and were, therefore, resistant to erythromycin, clindamycin, and quinupristin–dalfopristin.6 All isolates were susceptible to gentamicin and sulfamethoxazole-trimethoprim (Table 1). Of the 61 S. aureus isolates, 11 (18.0%) were considered MDR.9
Resistance profile of S. aureus isolates obtained from tonsils of patients submitted to tonsillectomy.
| Antimicrobial | Resistance | |
|---|---|---|
| n | % | |
| Penicillin | 51 | 83.6 |
| Cefoxitin | 16 | 26.2 |
| Ciprofloxacin | 15 | 24.6 |
| Erythromycin | 10 | 16.4 |
| Amoxicillin-clavulanate | 08 | 13.1 |
| Tetracycline | 07 | 11.5 |
| Clindamycin | 06 | 9.8 |
| Q/D | 06 | 9.8 |
| Ceftriaxone | 03 | 4.9 |
| Rifampin | 01 | 1.6 |
| Linezolid | 01 | 1.6 |
| Gentamicin | 00 | 0.0 |
| Trimethoprim-sulfamethoxazole | 00 | 0.0 |
According to the CLSI,8 cefoxitin is used to predict oxacillin resistance mediated by mecA gene expression. In the present study, 26.2% (16/61) of the S. aureus isolates were resistant to cefoxitin and the mecA gene was detected in two isolates. The gene coding for PVL was found in one MDR isolate, susceptible to methicillin.
The isolates showed high genetic diversity according to the similarity analysis performed by PFGE. Only Clusters 5 and 8 grouped genetically identical isolates from different patients. In nine patients (ID: 17, 36, 85, 90, 111, 124, 129, 133, 134), more than one genetically different isolate was identified colonizing the tonsil, and in one of them (ID: 133), four different isolates were simultaneously obtained (Fig. 1). Cluster 5 grouped nine isolates presenting more than 80% of similarity, with two clonal pulsotypes, obtained from different patients (Fig. 1).
No clonal relationship was observed between the two MRSA isolates, which shows that these phenotypes are dispersed among the different isolates obtained from the tonsils.
DiscussionBacterial pharyngotonsillitis is an infection that affects mainly children and adolescents between five and 15 years old.13,14 In this study, the mean age of the patients involved was 11.3 years, in line with the literature data. During childhood, due to the various contacts of children at home, school, and day care centers, there is an increase in variation of oral microbiota, leading to an increase in the frequency of these infections.15 There was no significant sex difference among those undergoing tonsillectomy, as observed in other studies.14
Recurrent pharyngotonsillitis was reported in 75.5% of the patients, showing its importance for the indication of tonsillectomy. According to some studies, recurrent pharyngotonsillitis and/or tonsillar hypertrophy, with sleep apnea, nocturnal snoring and respiratory pauses are the main indications for tonsillectomy.13
The high prevalence of S. aureus found (40.7%) in this study indicates bacterial persistence in the tonsils after an inflammatory process and treatment with antimicrobials. The throat region and the anterior nostril region are considered the primary colonization sites of S. aureus.16 The identification of S. aureus as the main agent of tonsillitis or as a probable co-pathogen has been reported by several authors, with prevalence as high as 83.0%.3,4,17,18
The presence of S. aureus in tonsil infections and its persistence in tonsillar tissue even after the inflammatory process may be related to its ability to form biofilm. The presence of the biofilm can explain therapeutic failures and, therefore, recurrence of the infection, and represent an important element of chronicity, even in the absence of acute inflammatory process.3
The resistance of the isolates to penicillin was 83.6% and 13.1% to the association of amoxicillin and clavulanate. This is evidence that the high rate of resistance found was due to β-lactamase enzyme production, which limits the use of this common therapeutic option in clinical practice.19
The therapeutic failures observed with penicillin led to an increase in the use of other antimicrobials, such as the association with β-lactamase inhibitors and cephalosporins.19
Another concern regarding resistance profiles of S. aureus is the emergence of MRSA strains, both in the community and in the hospital environment.6,20 In this study, two isolates (3.3%) were considered MRSA, in line with the results of Zautner et al.4 who found isolation of MRSA in just one out of 76 isolates (1.3%) from patients with recurrent tonsillitis in Germany. In Japan, Hirakata et al.21 identified 9.1% of MRSA isolates in patients with pharyngotonsillitis symptoms, and in the USA, Brook & Foote22 found 16.0% of MRSA in tonsils of patients submitted to tonsillectomy due to recurring tonsillitis. These data show that there seems to be no relationship between MRSA and pharyngotonsillitis under the conditions of our study, considering that only outpatients were included.
Although cefoxitin resistance is the best marker for MRSA screening,8 only two of the 16 isolates (12.5%) resistant to cefoxitin were identified as MRSA by detection of the mecA gene. In these cases, methicillin resistance may be due to mutation in genes encoding normal PBP or by overproduction of β-lactamases.23
Mutations in genes coding for PBPs can generate structural modifications, which alter the binding of these proteins to β-lactams antibiotics by decreasing their affinity and determining antimicrobial resistance.24 Mutations can also lead to overexpression of PBPs and produce a small but significant increase in resistance to β-lactam antibiotics. These mutations can be generated by the selective pressure exerted by excessive use of β-lactams.25
In addition to the changes in PBPs, S. aureus may develop resistance to β-lactams due to hyperproduction of β-lactamases.23 A study of McDougal and Thornsberry26 shows that S. aureus producing large amounts of β-lactamases can also inactivate more slowly penicillin resistant to penicillinases. In this study, the production of large amounts of β-lactamases may have influenced the cefoxitin disk test and determined phenotypic resistance.
Another possibility is the existence of a gene homologous to the mecA gene, the mecA LGA251, or mecC gene. This gene has 63% homology at the amino acid level and 70% at the DNA level with the mecA gene encoding the PBP2a protein.27 So far, isolates with the mecC gene have shown phenotypic resistance to β-lactams, but have not been identified in conventional PCRs for the mecA gene.8 An explanation for this phenomenon would be that PBP2a encoded by the mecC gene could have relatively high affinity to oxacillin but a low affinity to cefoxitin.28
The resistance rate of 24.6% to ciprofloxacin is worrisome. This drug has been reported to be effective against several pathogenic agents of tonsillitis, including S. aureus, and increased resistance leaves few therapeutic options to treat this condition.29
Inducible MLSb resistance profile was detected in 9.8% of the isolates. This resistance may be mediated by the presence of erm gene, which produces modification at the drug binding site in rRNA and causes therapeutic failures and relapses.30 Clindamycin is a widely used therapeutic alternative for staphylococcal infections, well tolerated by children, and the most frequent option for patients allergic to penicillin.31 Therefore, the iMLSB profile (inducible resistance to clindamycin) should be considered, when performing the susceptibility laboratory tests, in order to prescribe the adequate antimicrobial therapy to the patient.
One of the major concerns when analyzing resistance profiles of S. aureus is the presence of MDR strains. In this study, 11 (18.0%) MDR isolates were identified. The patients involved in the study were treated as outpatients. Mainly because they are children and adolescents who are in constant contact with other children and their relatives, the spread of MDR can overcome hospital barriers, making therapy and dissemination of these strains a challenge.18
It is noteworthy that one isolate presented the gene coding for PVL. This is an important virulence factor of S. aureus that causes serious infections such as necrotizing pneumonia. PVL-positive S. aureus are more associated with children and young adults, and may carry different lysogenic phage that contain genes for PVL. These can be easily transmitted horizontally and infect other PVL-negative strains of S. aureus.32,33
The similarity analysis of the isolates demonstrated genetic diversity between them. The presence of multiple isolates in the same patient favors the transfer of genetic information between them.34 This demonstrates the dynamics of colonization and the possibility of genetic alteration of the isolates that persist colonizing the tonsil, even in clinically healthy patients.
This diversity of genotypic profiles was also observed by Zautner et al.,4 where 76 S. aureus isolates were grouped into 24 different profiles. This demonstrates that under the study conditions, there is no clone associated with infections and that isolates are dispersed in this population. Probably, the factors associated with colonization and persistence of S. aureus in the tonsils are common in the species.
The clustering of similar isolates from different patients suggests that these may have a common origin and are associated with the pathogenesis of recurrent pharyngotonsillitis. Future investigations are necessary to determine the common genetic characteristics of these isolates that lead to dissemination in patients with the same clinical profile.
The results point to a change in the paradigm of diagnosis and treatment of recurrent pharyngotonsillitis in order to allow the correct use of antimicrobials and to reduce recurrence, which is the main cause of tonsillectomy.
FundingTo National Council for Scientific and Technological Development (CNPq) for financial support
Conflicts of interestThe authors declare no conflicts of interest.
To National Council for Scientific and Technological Development (CNPq) for financial support.