The aim of this study is to report the occurrence of the first outbreak of food poisoning caused by Salmonella Alachua in Brazil, as well as the antimicrobial susceptibility and the genetic relatedness of Salmonella Alachua strains isolated from clinical and food samples.
Material and methodsTo elucidate the outbreak, an epidemiological investigation was carried out, and two samples of common food were tested – mayonnaise salad and galinhada (a traditional Brazilian dish of chicken and rice) – according to the Compendium of methods for the microbiological examination of foods. Five stool samples were tested employing classic methods for the isolation and identification of enterobacteria. Strains of Salmonella were characterized for antibiotic susceptibility according to the Clinical and Laboratory Standards Institute guidelines (2013), and submitted to pulsed-field gel electrophoresis analysis, performed according to the Centers for Disease Control and Prevention PulseNet protocol.
ResultsA total of 94 people were interviewed after ingesting the food, 66 of whom had become ill. A 60-year old female patient who was hospitalized in a serious condition, developed septic shock and died two days after consuming the food. The presence of Salmonella Alachua was confirmed in all the analyzed stool samples, and in the two types of food. The five strains showed higher than minimum inhibitory concentration values of nalidixic acid (≥256μg/mL) and reduced ciprofloxacin susceptibility (minimum inhibitory concentration=0.5μg/mL). The pulsed-field gel electrophoresis analysis revealed indistinguishable patterns in Salmonella Alachua strains isolated from clinical and food samples.
ConclusionThe data presented herein confirm the foodborne disease outbreak. They also allowed for the identification of the source of infection, and suggest that products from poultry are potential reservoirs for this serotype, reinforcing the importance of warning consumers about the danger of possible contamination.
Salmonella, a genus of zoonotic enterobacteria responsible for outbreaks of infections in both humans and animals, has significant economic importance worldwide.1–3 It is estimated that Salmonella causes 93.8 million human infections and 155,000 deaths per year around the world.3 From 2000 to 2013, Salmonella was the infectious agent most commonly linked to outbreaks of food poisoning in Brazil, with a total of 1560 episodes representing 38.3% of all agents identified during the said period.4
Both children and the elderly, as well as immunocompromised individuals with salmonellosis may see the condition evolve to more severe stages as, upon entering the bloodstream, the bacteria can cause extraintestinal infections.5 The risk of invasive disease is two to six times higher than with other foodborne pathogens6; the death rate is also higher.7
Salmonella Alachua was first described in 1952, during a study of the effects of salmonellosis in pigs in the city of Alachua, Florida, where it was isolated in a soil sample from a pig farm.8 However, the first reported isolation in animals was in 1955 after several outbreaks of enteritis in chickens from different farms in Bombay, India, that resulted in a considerable loss of birds.9 Since then, isolation has proved uncommon worldwide, as it is found in human and non-human samples at a rate of from 0.03 to 3.8%.10–14
In this study, we report the occurrence of the first outbreak of food poisoning caused by Salmonella Alachua in Brazil, in a city in the northwestern region of the State of São Paulo. Moreover we report of the antimicrobial susceptibility and the genetic relatedness of Salmonella Alachua strains isolated from clinical and food samples.
Materials and methodsEpidemiological investigationIn order to better explain the occurrence of a foodborne disease outbreak in a city in the northwestern region of the State of São Paulo in November 2012, an epidemiological investigation was carried out by the local health surveillance team, with the collection of samples of the ingested food – mayonnaise salad and galinhada (a traditional Brazilian dish of chicken and rice) – and stool samples from five patients. All the tests were performed at the Regional Laboratory Center of Instituto Adolfo Lutz in São José do Rio Preto (RLCIAL).
Microbiological analysis of the foodThe food was analyzed according to the methods described in the Compendium of Methods for the Microbiological Examination of Foods – APHA15 for contamination by coliform group bacteria, Staphylococcus aureus, Bacillus cereus, Clostridium perfringens and Salmonella.
The procedures for isolation and identification of Salmonella, were carried out through pre-enrichment of 25g samples by homogenization with 225mL lactose broth (10−1 dilution), and incubation overnight at 36±1°C. Selective enrichment was performed in tetrathionate (TT) broth and modified Rappaport-Vassiliadis (RV) broth, followed by incubation at 36±1°C for 24h and 42°C for 24–48h, respectively. Each enrichment broth was streaked onto selective plates: Salmonella-Shigella agar (SS), brilliant green agar (BG) and xylose lysine deoxycholate agar (XLD), and incubated for 24h at 36±1°C.15
Even though no other biochemical tests were performed, characteristic colonies of each plate were biochemically tested using only IAL medium16 for the presumptive identification of Enterobacteriaceae and incubated for 24h at 36±1°C. Strains with presumptive identification of Salmonella were submitted to serological tests using polyvalent somatic (O) and flagellar (H) antisera produced by the Laboratory of Enteric Pathogens of the Instituto Adolfo Lutz.
The standard methodology for the study of Salmonella15 recommends the use of the presence/absence method in 25g of a food sample. The highest dilution in which Salmonella is demonstrably present in the food sample was used as a complement to the testing, with the purpose of determining which food had the highest microbial load. Albeit important, this is not a quantitative method. For this, we started with 10mL of a 10−1 dilution, serial dilutions of the food samples were performed in tubes containing lactose broth up to a dilution of 10−9. After incubating the tubes for 18–24h at 36±1°C, the presence of turbidity was verified in the different dilutions. The inoculum from all tubes that presented turbidity was submitted to selective enrichment, isolation and identification procedures, pursuant to the APHA methodology.15
Stool analysisStool samples were collected from a total of five patients by swab and transported in Cary-Blair medium to investigate Escherichia coli, Aeromonas spp., Shigella spp. and Salmonella spp. At RLCIAL, the swabs were seeded in plates with MacConkey Agar (MC), Salmonella-Shigella Agar (SS) and Sorbitol MacConkey Agar (MCS), and incubated for 24h at 36±1°C. Subsequently, the swab was placed in a tube containing 10mL of Tetrathionate (TT) broth and incubated for 24h at 36±1°C for selective enrichment. After this period, the TT broth was inoculated on plates with MC and Brilliant Green Agar. After incubation for 18–24h at 36±1°C, all the plates with the selective medium were examined for colony morphology and for utilization of lactose/sorbitol, which are used to inoculate the IAL medium16 for the presumptive identification of the researched microorganisms.
Strains with presumptive identification of Salmonella were submitted to serological tests using polyvalent somatic (O) and flagellar (H) antisera produced by the Laboratory of Enteric Pathogens, Instituto Adolfo Lutz.
SerotypingAll the isolates of Salmonella from the food and stool samples were sent to the Central Laboratory of the Instituto Adolfo Lutz (CLIAL) for complete serotyping on the basis of somatic O and phase 1 and phase 2 of the H flagellar antigens by agglutination tests with antisera prepared in the Laboratory of Enteric Pathogens, Institute Adolfo Lutz, São Paulo as specified in the Kauffmann–White protocol for Salmonella serotyping.17
Susceptibility testingAntimicrobial susceptibility testing was performed for all isolates using the disk diffusion method according to the guidelines of the Clinical and Laboratory Standards Institute – CLSI.18 The following antimicrobial disks (Oxoid) were used: nalidixic acid (30μg), amoxicillin–clavulanic acid (20/10μg), amikacin (30μg), ampicillin (10μg), aztreonam (30μg), ceftazidime (30μg), cefotaxime (30μg), ceftriaxone (30μg), cefepime (30μg), ciprofloxacin (5μg), chloramphenicol (30μg), streptomycin (10μg), gentamicin (10μg), imipenem (10μg), trimethoprim–sulfamethoxazole (1.25/23.75μg), sulfonamide (250μg), and tetracycline (30μg). Categorization of the diameter of halos in susceptible, intermediate or resistant followed CLSI recommendations.18
Minimum inhibitory concentrations (MIC) were determined for nalidixic acid and ciprofloxacin by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations. The range of MIC of ciprofloxacin for Salmonella was recently changed to susceptible: ≤0.06μg/mL; intermediate susceptible: 0.12–0.5μg/mL; resistant: ≥1μg/mL.18
E. coli ATCC 25922 and E. coli ATCC 35218 were used as reference strains for antimicrobial susceptibility testing.
Pulsed field gel electrophoresisPulsed field gel electrophoresis (PFGE) analysis was performed for all the isolates at CLIAL according to the Centers for Disease Control and Prevention (CDC) PulseNet protocol (www.cdc.gov/pulsenet/pathogens/index.html). Briefly, cell lysis was followed by proteinase K treatment and DNA restriction with XbaI (New England Biolabs, Ipswich, MA). Electrophoresis was performed with a CHEF DRIII system (BioRad Laboratories Inc., Hercules, CA) using the following run parameters: a switch time of 2.2–63.8s and a run time of 20h. Salmonella Braenderup H9812 was used as a molecular size marker.19 TIFF images were analyzed using the BioNumerics 5.0 software (Applied Maths). Dice's coefficient with tolerance of 1.5 was used to calculate similarity using the Unweighted Pair Group Method with arithmetic averages (UPGMA).
ResultsEpidemiological investigationOf the 94 people interviewed after the foodborne outbreak, the epidemiological investigation found that the consumption of mayonnaise salad and galinhada was common to the entire group; 66, both children and adults, had become ill. The median incubation period was 72h, and the main symptoms observed were: diarrhea 63/66 (95.4%), abdominal pain 50/66 (75.7%), nausea 40/66 (60.6%), fever 27/66 (40.9%), vomiting 23/66 (34.8), and headache 22/66 (33.3%). Attack rates by age group are shown in Table 1.
Attack rate by age group of the subjects exposed to risk by the ingestion of food contaminated by Salmonella.
| Age range (years) | Subjects exposed to risk | Total, n | Attack rate (%) | |
|---|---|---|---|---|
| Individuals sick, n | Individuals not sick, n | |||
| <1 | – | – | – | – |
| 1–9 | 7 | 1 | 8 | 87.5 |
| 10–20 | 6 | 2 | 8 | 75.0 |
| 20–39 | 28 | 11 | 39 | 71.8 |
| 40–49 | 10 | 4 | 14 | 71.4 |
| 50–59 | 5 | 7 | 12 | 41.7 |
| ≥60 | 10 | 3 | 13 | 76.9 |
| Total | 66 | 28 | 94 | 70.2 |
According to the investigation, a 60-year old female patient who was hospitalized in a serious condition, developed septic shock and died two days after consuming the food.
Microbiological analysisThe presence of Salmonella was confirmed in all the analyzed stool samples and in both types of food, consequently it was isolated in dilutions of 10−7 and 10−2 of the salad mayonnaise and galinhada, respectively. No other pathogens were isolated from the food or stool samples.
The Most Probable Numbers (MPN) of thermotolerant coliforms found in the mayonnaise salad and galinhada samples were >2400/g and 240/g, respectively.
SerotypingAll the strains isolated from human and food sources were identified as Salmonella enterica serovar Alachua by agglutination tests.
Antimicrobial susceptibilityAll the Salmonella Alachua strains demonstrated resistance to nalidixic acid and reduced susceptibility to ciprofloxacin (intermediate resistant). However, all of them were susceptible to the other antimicrobials tested.
The seven strains showed higher MIC values for nalidixic acid (≥256μg/mL) and reduced ciprofloxacin susceptibility (MIC=0.5μg/mL).
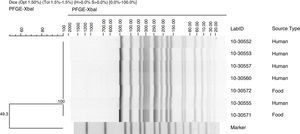
Pulsed field gel electrophoresisA dendrogram, generated by PFGE patterns of Salmonella Alachua strains using XbaI as the restriction enzyme, is shown in Fig. 1. One PFGE pattern was identified among the Salmonella Alachua clinical and food isolates analyzed. The genetic relatedness among the strains was 100%.
DiscussionCurrently, Salmonella is one of the most common microorganisms involved in foodborne disease outbreaks worldwide.4,20,21 In the United States, Salmonella Alachua corresponds to 0.05% of the isolates identified in the period from 1999 to 2009, while Salmonella Typhimurium and Salmonella Enteritidis were the prevalent serotypes, accounting for 18.5% and 16.3% of cases, respectively.13
A similar situation occurred in Mexico, where, between 1972 and 1999, only 26 (0.1%) strains of 24,394 Salmonella isolates from various public health and private laboratories were found to be Salmonella Alachua.12
Three (0.03%) and one (0.04%) isolates of Salmonella Alachua identified in non-human and human material, respectively, were registered in the State of São Paulo, Brazil, in different periods.11,22 Moreover, in the State of Goiás, Brazil, Salmonella Alachua was isolated in two (3.8%) samples from bird transport box liners.14
According to Almeida et al.23 no presence of Salmonella Alachua was observed in human and non-human (food) matter during the 1990s in the same region as the current reported outbreak. The only time in which this serotype was isolated was in 2007, from a sample of raw eggs, during an investigation of a foodborne disease outbreak. However, it was not considered the causative agent, as the Salmonella enterica serotype Infantis was isolated in all the stool samples of the affected individuals (12 patients).24
A significant increase in the number of Salmonella Alachua isolates (27 to 88) observed in the USA in 1982, corresponding to an upsurge of 226% over the previous year, was attributed to the adoption of children from a nursery in Calcutta, India, by American families.25 Given the above, one should consider the possibility that this serotype may have had a relevant epidemiological expression for some time in India.
Changes in the prevalence of serotypes have been observed in several studies,22,26–28 hence any serotype, however unusual or uncommon, may become emergent and cause serious infections or outbreaks.
Some serotypes are frequently associated with certain classes of food. Thus, studies on the serotypes characterization provide information on reservoirs, routes of transmission and prevalence in a specific region, particularly when outbreaks of foodborne diseases occur.4,29,30
Notwithstanding the fact that there are few reports on Salmonella Alachua, products originating from poultry farms can be considered possible reservoirs for this serotype.14,24
Considering the fact that Salmonella Alachua has been identified with the same genetic connection in both the isolated foods, it can be suggested that cross-contamination occurred between the two types of food analyzed. Cross-contamination can occur as a result of inadequate manipulation, and use of contaminated kitchen utensils, and may become critical, depending on the amount of time that the product is exposed to improper storage temperatures.31 It should be emphasized that the use of raw eggs in the preparation of the mayonnaise salad during the epidemiological investigation of the outbreak was not confirmed. Therefore, the chicken meat used for the preparation of galinhada can be considered the likely source of Salmonella Alachua.
The infectious dose of Salmonella varies between 105 and 108 cells, with infective doses as low as ≤103 being reported in immunocompromised patients, while certain serotypes are related to foodborne disease outbreaks.5,32 Consequently, despite the non-quantification of Salmonella, the large number of affected individuals in this study can be explained by the presence of this pathogen at dilutions of 10−7 in the sample of mayonnaise salad.
Certain reports have demonstrated antimicrobial resistance to Salmonella Alachua strains from both human and non-human sources.14,33,34 In this study, even though there was susceptibility of Salmonella Alachua strains to most of the antimicrobials tests, all presented resistance to nalidixic acid (MIC ≥256μg/mL) with reduced susceptibility to ciprofloxacin (MIC=0.5μg/mL). The resistance to nalidixic acid can predict a resistance to fluoroquinolones, as observed in the study.
Fluoroquinolones are currently used to treat invasive and systematic salmonellosis, occurring in humans. These are also effective in treating a range of different infections encountered in animals. Resistance to fluoroquinolones is relatively uncommon with Salmonella. However, in recent years, studies have reported an increase in the number of clinical isolates with reduced susceptibility to ciprofloxacin associated with treatment failure.35–37 The emergence of reduced susceptibility to fluoroquinolones among food animals and humans is considered a significant public health concern, and should be carefully monitored.
PFGE revealed indistinguishable patterns in Salmonella Alachua strains isolated from clinical and food samples, thus confirming the foodborne disease outbreak; this also allowed for the identification of the source of infection. PFGE is a standard typing method used in Salmonella outbreak investigations to determine the relationship and distribution of genetic subtypes of Salmonella circulating in countries, as well as the application for the investigation of foodborne outbreaks, and to detect emerging pathogens.38
ConclusionThis study reports on the first foodborne disease outbreak caused by the Salmonella Alachua serotype in Brazil. The source of infection was confirmed by PFGE, and all Salmonella Alachua strains presented resistance to nalidixic acid, and reduced susceptibility to ciprofloxacin.
The findings of this study highlight the importance of the numerous and complex activities of Public Health Laboratories in the development of necessary knowledge to optimize prevention and food contamination control.
Conflicts of interestThe authors declare no conflicts of interest.