The aim of this study was to evaluate the use of RNA interference to inhibit herpes simplex virus type-1 replication in vitro. For herpes simplex virus type-1 gene silencing, three different small interfering RNAs (siRNAs) targeting the herpes simplex virus type-1 UL39 gene (sequence si-UL 39-1, si-UL 39-2, and si-UL 39-3) were used, which encode the large subunit of ribonucleotide reductase, an essential enzyme for DNA synthesis. Herpes simplex virus type-1 was isolated from saliva samples and mucocutaneous lesions from infected patients. All mucocutaneous lesions’ samples were positive for herpes simplex virus type-1 by real-time PCR and by virus isolation; all herpes simplex virus type-1 from saliva samples were positive by real-time PCR and 50% were positive by virus isolation. The levels of herpes simplex virus type-1 DNA remaining after siRNA treatment were assessed by real-time PCR, whose results demonstrated that the effect of siRNAs on gene expression depends on siRNA concentration. The three siRNA sequences used were able to inhibit viral replication, assessed by real-time PCR and plaque assays and among them, the sequence si-UL 39-1 was the most effective. This sequence inhibited 99% of herpes simplex virus type-1 replication. The results demonstrate that silencing herpes simplex virus type-1 UL39 expression by siRNAs effectively inhibits herpes simplex virus type-1 replication, suggesting that siRNA based antiviral strategy may be a potential therapeutic alternative.
Herpes simplex virus type 1 (HSV-1) is a member of the Herpesviridae family and is characterized by its ability to establish latency after primary infection and subsequently reactivated.1 Herpes simplex virus (HSV) is an enveloped, double-stranded (ds) DNA virus. The HSV-1 genome consists of 152kb of linear dsDNA arranged as long and short unique segments (UL and US) flanked by inverted repeated sequences (TRL/IRL and IRS/TRS, respectively).2 Worldwide prevalence of HSV ranges from 65% to 90%. HSV-1 gives rise to a spectrum of clinical manifestations and can still be a major cause of morbidity and mortality.3 HSV-1 is the causative agent of encephalitis, corneal blindness, and several peripheral nervous system disorders.4 Beyond the neonatal period, most childhood herpes simplex virus infections are caused by HSV-1. The seroprevalence of HSV-1 antibodies increases with age, reaching 20% by the age of five years. No increase occurs until 20–40 years of age, when 40–60% of individuals are HSV-1 seropositive. Mortality associated with herpes simplex virus is primarily related to perinatal infection, encephalitis, and infection in individuals who are immunocompromised.4,5
Recently, RNA interference (RNAi) has emerged as a new therapeutic strategy against viral infection.6 RNAi is now widely used to knockdown gene expression, in a sequence-specific manner.6 It can inhibit the expression of crucial viral proteins by targeting viral mRNAs for degradation instead of the proteins they encode.7 RNAi is mediated by 21–25 nucleotide double-stranded small interfering RNA (siRNA) molecules. siRNAs are incorporated into the RNA-induced silencing complex (RISC), which mediates mRNA sequence-specific binding and cleavage.8 In particular, siRNAs, processed from double-stranded (ds) RNA precursors by the type III endoribonuclease Dicer, mediate post-transcriptional gene silencing (PTGS).9 Some studies confirm that siRNA-directed transcriptional gene silencing is conserved in mammalian cells.10 Small RNAs may guide mammalian transcriptional silencing in many different biological contexts.10 HSV-1 encodes its own ribonucleotide reductase (RR), which reduces ribonucleoside diphosphates to the corresponding deoxyribonucleotides and is essential for replication. The HSV-1 RR is formed by a large subunit designated ICP6, encoded by the UL39 gene, and a small subunit which is encoded by the UL-40 gene. The HSV-1 cannot utilize cellular RR and therefore is dependent upon its own reductase for replication.11 siRNA-based antiviral therapy may be a potential effective therapeutic alternative for patients with acyclovir-resistant HSV strains.
This study evaluated the effects of siRNAs targeting the HSV-1 UL39 gene on the replication of HSV-1 isolated from mucocutaneous lesions and saliva samples. Infected patients with blisters and sores characteristic of herpes skin disease who had HSV-1 DNA detected were included in this study. For this purpose, samples were collected between 2009 and 2010 after obtaining informed consent statement from each individual. This study was approved by Ethics Committee of Oswaldo Cruz Foundation (protocol number: 544/09). The saliva samples were collected using ChemBio (Medford, New York) and samples from mucocutaneous lesions were collected using Salivette (Sarsdedt, Germany) devices, respectively. The detection of HSV-1 was confirmed by virus isolation in cell culture and real-time PCR. In brief, samples from mucocutaneous lesions or saliva samples were suspended in 3mL of medium 199 (Sigma) containing antibiotics and antifungical (2.5μg/mL of each one respectively). Vero cell cultures were inoculated with 300μL of solution containing mucocutaneous or saliva samples. The HSV-1 strain KOS was used as control of infection.12 During 15 days cell cultures were observed for viral cytopathic effect typical for HSV infection. HSV DNA was extracted from clinical specimens using commercial kits (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. HSV-1 specific PCR analysis was conducted with a SYBR Green real-time PCR assay according to manufacturer's instructions. The real-time quantitative PCR was performed with oligonucleotide primer pairs specific for the coding region of the glycoprotein D (gD) of HSV-1, as reported previously.13 The primers used were HSV-FP (5′-CGGCCGTGTGACACTATCG-3′) and HSV-RP (5′-CTCGTAAAATGGCCCCTCC-3′).13 A standard curve was prepared by serial dilution (101–107) from DNA extracted from KOS strain (108copies/mL).
After confirming HSV1 infection, the replication of HSV-1 was inhibited using three siRNA molecules against the UL-39 gene from HSV-1 (si-UL 39-1, si-UL 39-2, si-UL39-3).14 One sequence not targeting any known gene was used as a negative control of siRNA (Applied Biosystems, Foster City, CA, USA). Vero cells were grown in 6-well plates to 80%–90% confluence and then transfected with specific or control siRNA using the commercial kit siPORT™ Amine (Ambion/Applied Biosystems, Foster city, CA, USA). After four hours, the cells were infected with HSV-1 from the infected patients (25 PFU/mL). At 48–72h post-infection, plates were fixed with 10% paraformaldehyde for two minutes and then stained with 1% crystal violet for 30minutes to count the number of plaques per well. The effects of siRNA in infected cells were also measured by quantification of HSV DNA. After 48h, the DNA from the infected cells was extracted using a commercial kit (Qiagen) following the manufacturer's instructions. Real-time PCR relative quantitative reactions were performed using SYBR Green real-time PCR Master Mix (Roche, New Jersey, USA) and 18S RNA was used as the endogenous control. The statistical analysis was performed using the programs “Graph Pad Prism” 5.0 and Excel. The data were reported as mean±standard deviation (SD) and the levels of significance were evaluated using the Student's t-test and ANOVA. Differences were considered significant when p<0.05.
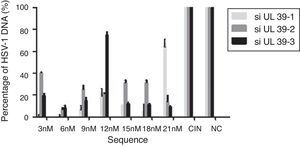
All samples of mucocutaneous lesions were positive for HSV-1 by real time-PCR and by virus isolation. The viral load in mucocutaneous lesions samples ranged from 3.85×103 to 9.78×104copies/mL. The saliva samples were all positive for HSV-1 by real-time PCR, and only 50% of samples were positive by viral isolation. The virus load in saliva samples ranged from 2.44×103 to 1.54×104copies/mL. Previous studies have shown that the isolation of HSV DNA from saliva, using the method of viral isolation, is hampered by the presence of substances with anti-HSV activity in saliva.15,16 Using the highly sensitive technique of real-time PCR, we detected the HSV-1 even in those samples with low viral load. The concentration of siRNA is important to suppress virus replication. Aiming to evaluate the best concentration of siRNA required to suppress HSV replication, cells were transfected with siRNAs in different concentrations (final concentration 3nM, 6nM, 9nM, 12nM, 15nM, 18nM, 21nM). In this experiment two controls were used: (1) cells infected with HSV-1 that were transfected with non-specific siRNA (NC) and cells infected with HSV-1 and not transfected (CIN).
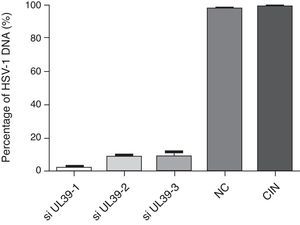
The results demonstrated that these siRNAs could potently inhibit HSV-1 replication in vitro and was observed that the concentration of UL39 specific siRNAs to achieve the highest inhibition of HSV transcription was 6nM (Fig. 1). This concentration is considerably less than those used in other studies of gene silencing in mammalian cells, which has typically ranged from 20 to 200nM.17–19 However, low concentrations around 10nM have also been shown to be sufficient for an effective silencing of genes.20,21 Following the establishment of the concentration of siRNA that had the highest inhibitory effect on UL-39 transcription, a sample from mucocutaneous lesions with the highest viral load (9.78×104copies/mL) was used to evaluate the efficiency of gene silencing. The concentration of 6nM of siRNAs was used for transfection and gene silencing was evaluated 48hours post-infection. Analysis by real-time PCR showed that 99, 90 and 88.5% of gene silencing were achieved upon transfection with siRNAs targeting si-UL 39-1, si-UL39-2, and si-UL 39-3 sequences, respectively (Fig. 2). In our study, the siRNA specific to the UL-39-1 sequence showed the highest level of silencing. By plaque assays from 63 to 69% inhibition of viral plaque formation upon silencing the UL-39 gene. Previous study demonstrated that the inhibition rates of siRNA1 and siRNA2 on HSV-1 plaque formation were 35.51 and 51.62%.14 The differences in the level of silencing determined by these siRNA sequences may be due to the concentration of siRNA, conditions of transfection, viral strains, type of cell used and differences in the thermodynamic properties of the siRNAs. Currently, there has been increasing number of studies exploring the potential for RNAi approaches to HSV-1. The siRNAi used in this study silenced specifically the HSV-1 UL39 gene, which encodes the large subunit of ribonucleotide reductase, ICP6.14,22 RNAi has also been reported to inhibit HSV-1 replication by using siRNAs targeting glycoprotein E that plays key role in cell-to-cell spread and virus-induced cell fusion20; DNA polymerase gene and VP16 play vital roles in initiation of viral gene expression and viral proliferation22 and ICP4 is a major regulatory gene required for efficient transcription of early and late viral genes making it essential for lytic infection.19 These studies that applied RNAi to interfere HSV-1 infection suggested that these small sequences might have the potential for effective therapeutic alternative in patients with HSV-1 infection. One important aspect of using siRNA activity against DNA viruses is the need to show the inhibition of viral DNA replication, and when the amounts of viral DNA were quantified almost no viral DNA could be detected, which demonstrated the inhibition of genome replication. Herein, the siRNAi were effective to inhibit replication of HSV-1 in a strain adapted in cell culture (KOS) and also in wild-type virus isolated directly from an infected patient with high viral load. The inhibitory effects were related to the concentration of siRNAi transfected, thus determining the right concentration siRNAi can improve the inhibition of virus replication.
Effect of different siRNAs on the expression levels of HSV-1 DNA. The expression level of HSV-1 DNA in cells treated with different concentrations (3–21nM) of siRNAs (si-UL-39 1, si-UL-39 2, si-UL39 3) was determined by real-time quantitative PCR. The result was normalized to the housekeeping gene 18S RNA. NC – cells transfected with non-specific siRNA (negative control). CIN cells infected with HSV-1 but not transfected.
The authors declare no conflicts of interest.
National Council of Technological and Scientific Development (CNPq), 478979/2009-6 and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), E-26/111.561/2010 were acknowledged for financial support.