Human cytomegalovirus is a ubiquitous pathogen that infects the majority of the world's population. After long period of time co-evolving with human being, this pathogen has developed several strategies to evade host immune surveillance. One of the major trick is encoding homologous to those of the host organism or stealing host cellular genes that have significant functions in immune system. To date, we have found several viral immune analogous which include G protein coupled receptor, class I major histocompatibility complex and chemokine. Chemokine is a small group of molecules which is defined by the presence of four cysteines in highly conserved region. The four kinds of chemokines (C, CC, CXC, and CX3C) are classified based on the arrangement of 1 or 2 N-terminal cysteine residues. UL128 protein is one of the analogous that encoded by human cytomegalovirus that has similar amino acid sequences to the human CC chemokine. It has been proved to be one of the essential particles that involved in human cytomegalovirus entry into epithelial/endothelial cells as well as macrophages. It is also the target of potent neutralizing antibodies in human cytomegalovirus-seropositive individuals.
We had demonstrated the chemotactic trait of UL128 protein in our previous study. Recombinant UL128 in vitro has the ability to attract monocytes to the infection region and enhances peripheral blood mononuclear cell proliferation by activating the MAPK/ERK signaling pathway. However, the way that this viral encoded chemokine interacting with peripheral blood mononuclear cells and the detailed mechanism that involving the virus entry into host cells keeps unknown. Here we performed in vitro investigation into the effects of UL128 protein on peripheral blood mononuclear cell's activation and receptor binding, which may help us further understand the immunomodulatory function of UL128 protein as well as human cytomegalovirus diffusion mechanism.
Human cytomegalovirus (HCMV) is a ubiquitous pathogen that infects the majority of the world population.1 The infection caused by HCMV in the human body can be lifelong. HCMV has developed several strategies to evade host immune surveillance over a long period time of co-evolution with human beings. The major tricks are to encode proteins that are homologous to those of the host organism or to steal host cellular genes that has significant functions in the immune system.2 The potential roles of these immune analogous in virus trafficking, persistence, and immune evasion remain largely unknown.
To date, we have found several viral immune analogous which include G protein coupled receptor, class I major histocompatibility complex (MHC-I) and chemokines.3 Chemokines are a small group of molecules, which are defined by the presence of four cysteines in highly conserved region. The four kinds of chemokines (C, CC, CXC, and CX3C) are classified based on the arrangement of 1 or 2 N-terminal cysteine residues.4 UL128 protein is one of the analogous encoded by HCMV, which has similar amino acid sequences to the human CC chemokine. It proved to be one of the essential particles involved in HCMV entry into epithelial/endothelial cells as well as macrophages.5 UL128 protein is also the target of potent neutralizing antibodies (NAb) in HCMV-seropositive individuals.6
We had demonstrated the chemotactic trait of UL128 protein in a previous study.7 Recombinant UL128 in vitro has the ability to attract monocytes to the infection site and enhances peripheral blood mononuclear cells (PBMC) proliferation by activating the MAPK/ERK signaling pathway, which may contribute to virus dissemination from one infectious organ to other susceptible organs.8 However, the way that this viral encoded chemokine interacting with PBMC and the detailed mechanism involved in virus entry into host cells needs further study. Here we performed an in vitro investigation of the effects of UL128 protein on PBMC activation and receptor binding, which may further strengthen the immunomodulatory function of UL128 protein and contribute to better understand HCMV diffusion mechanism.
Material and methodsExpression and purification of recombinant UL128 proteinRecombinant UL128 protein was obtained as reported previously.7 The UL128 gene sequence position 1243-2003 was selected from the Genbank (Accession no: GU574790.1). UL128 gene product was cloned using the pIRES-AcGFP vector (Qiagen, Valencia, CA) and was employed for the bacterial expression of pIRES-AcGFP with a 6-Histag fused to its N-terminus. Chinese hamster ovary cells were used for production of recombinant UL128 protein and the target protein was obtained through His·Bind Kit (Novagen, Germany). Protein concentration was quantified using the Bradford assay (Biorad) according to the manufacturer's instruction.
Western blotSince recombinant UL128 protein was labeled by 6-Histag, western blot analysis was carried out using 6×His Rabbit Polyclonal Antibody (Epitomics, Cat.: S2917) to confirm the presence of UL128 protein. The purified recombinant UL128-histag from SDS PAGE was transferred to PVDF (polyvinylidene difluoride) membrane. The membrane was blocked in 5% defatted milk dissolved in PBST (PBS containing 0.5% Tween-20) with gentle shaking at room temperature for 2h. The membrane was incubated in a 1:1000 dilution of rabbit anti-his-polyclonal antibody in blocking buffer with gentle shaking at room temperature for 1h, then changed to 4°C overnight. After washing with PBST three times for 10min each, the membrane was incubated in a 1:2000 dilution of sheet anti-rabbit IgG horseradish peroxidase (HRP) conjugated as secondary antibody (Epitomics, Cat.: 3053-1) in blocking buffer, with gentle shaking at room temperature for 1h after the membrane was washed with PBST for three times.
Silver stainSilver stain was applied to detect trace amounts of proteins in gels. (1) Fixed: after electrophoresis (see above of SDS-PAGE), the gel was put in 100mL fixative liquid and shook for 20min at room temperature, placing overnight to further reduce background; (2) Washing: remove the from the fixed liquid, add 100mL of 30% ethanol and shake for 10min at room temperature. Discard ethanol, add 200mL double distilled water and shake for 10min at room temperature; (3) Sensitization: remove water, then add 100mL silver staining sensitizing solution (1×), shake for 2min at room temperature at a shaking speed of 60–70rpm (revolutions per minute); (4) Silver staining: add 100mL silver solution (1×), shake for 10min at a shaking speed of 60–70rpm, and washed by double distilled water for 1min; (5) Color: discard water, add 100mL silver staining solution, shake for 3–10min until the ideal protein band appears; (6) Termination: remove from silver staining solution, add 100mL silver staining terminated liquid (1×), shake for 10min at room temperature, and washed by double distilled water for 2min.
Peripheral blood mononuclear cell (PBMC) culturePBMC from whole blood of healthy volunteers were isolated by standard density centrifugation (Ficoll Separation Solution: GIBCO, Germany). The mononuclear cells were collected and washed twice in phosphate-buffered saline (PBS, pH 7.2). By adjusting to 1×106cells/mL, the cells were re-suspended in MEM medium with 10% heated-activated fetal calf serum. Informed consents were provided before sample collection.
Measurement of calcium concentrationAs immune cells activation is countered by calcium fluxion, the intracellular calcium concentration in response to UL128 was monitored using fluo-3 AM (Calbiochem; La Jolla, CA USA) in PBMCs. The PBMC calcium level was determined by flow cytometry (Millipore, easyCyte8). Cells were pre-treated with or without 10μM fluo-3 AM (intracellular calcium chelator) for 1h, then washed with PBS and diluted to 5×106cells/mL in Hank's balanced salt solution containing Ca2+ and Mg2+ and 1% FBS. Different concentrations of U.L128 were added to 100nM and incubated at 37°C for 30min, and then analyzed by flow cytometry. Relative intra-cellular calcium levels were measured as the ratio of emissions at 490nm to the emissions at 400nm according to the manufacturer's instructions.
Cross-linking assayThe cross-linking assay was used to prove the ability of UL128 protein to bind the membrane receptor of PBMC and compare with the positive control MIP-1α. PBMC (1×107cells/mL) were suspended in MEM medium pretreated with recombinant UL128 (10ng/mL) or MIP-1α(10ng/mL) for 24h then made into slides. Slides were washed four times with D-hanks (100nmol/L) containing 0.5% BSA and fixed with 4% paraformaldehyde for 20min. Slides were incubated with either anti-UL128 antibody (1:200, supplied by HangZhou NeuroPeptide Biological Science and Technology Incorporation) or rabbit anti-MIP-1αantibody (Rockland, 109-401-315) or normal rabbit serum at room temperature for 45min. Slides were washed four times with D-hanks and incubated with biotin-conjugated goat anti-rabbit IgG (1:200) at room temperature for 30min. The results were observed under microscope by counting the positive cells. The experiment was repeated three times for data analysis.
Quantitative real-time PCR (qPCR)qPCR analyses were performed to quantify the CC receptor gene expression. Purified recombinant UL128 protein was co-cultured with PBMC for 24h before CC receptor detection. PBS added in PBMC was used as negative control and MIP-1α co-cultured PBMC was tested as positive control. Conserved regions at appropriate distances of CC receptors were used for designing the primers employed in qPCR reactions (Table 1). This was achieved using Primer Express 3.0 software. Total RNA was extracted using the RNeasy kit (Qiagen). RNA concentration and purity was estimated using a NanoVue Spectrophotometer and assessed for quality using agarose gel electrophoresis. Total RNA (2mg) was reverse transcribed to cDNA using the Fermentas First Strand Synthesis kit (Invitrogen). Amplifications were performed in an ABI Prism 7500 sequence detection system (Applied Biosystems). Fifteen μL reactions were prepared in clear 96 well fast plates. Amplification mixture concluded with the Maxima SYBR Green qPCR Master Mix (2×) (Thermo Scientific, USA), and a 0.2μM end concentration of each of the forward and reverse primers, 267nM Rox, and 25ng template cDNA. Amplification was carried out with a preliminary 2min UDG pretreatment at 50°C followed by a 10min initial denaturation at 95°C, followed by 40 cycles of denaturation for 15s at 95°C and annealing/extension for 60s at 60°C. Each experiment contains no template and no reverse transcriptase controls.
Primers of CC chemokine receptors.
| Gene | Locus | Primer sequences | Product size (bp) |
|---|---|---|---|
| CCR1 | Exon2 (61–481) | Forward: GACTATGACACGACCACAGAReverse: GTAGATGGAGGACTTGGACCGGTAA | 225 |
| CCR2 | Exon1 (1–434) | Forward: ACAGGAGCAGATGTACAGReverse: GTGATCATCCTCTCGTCTCT | 253 |
| CCR3 | Exon1 (238–780) | Forward: CAGGAGGCTCCGAATTATGAReverse: ACTATGGTCTCGTGACTACC | 180 |
| CCR4 | Exon2 (118–660) | Forward: CCACGGATATAGCAGACACCReverse: GAACTCCTCGGACATCTCAA | 450 |
| CCR5 | Exon2 (58–346) | Forward: GGCTGAAGAGCATGACTGACReverse: GATGGACGAGTTGGACCGGT | 403 |
Results from real-time PCR and FCM were derived from at least three independent experiments. Statistical significance between siRNA and control groups was evaluated with one-way ANOVA followed by the Dunnett post-test using the Prism3 GraphPad software. Significance level was calculated using an assigned confidence interval of 95%. p<0.05 was considered significant.
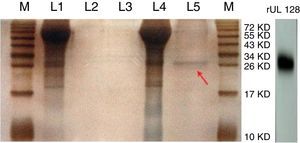
ResultsPurification of recombinant UL128 proteinThe rUL128 production system was successfully constructed by using eukaryotic expression vector (pIRES-AcGFP-UL128) with 6× His-Tag fused to its N-terminal, then transferred into Chinese Hamster Ovary (CHO) cell line. Purified rUL128 was about 26kilodalton (KD) and was condensed into 0.1mg/mL (Table 2). Silver stain experiment confirmed high purification of the destined protein (Fig. 1). As shown in the figure, the red arrow refers to purified recombinant UL128 protein, which was consistent with western blot analysis.
Concentration of purified UL128 protein.
| Standard (mg/mL) | Standard OD value (λ=570nm) | Sample OD value (UL128 purification) | Sample concentration (mg/mL) |
|---|---|---|---|
| 0 | 0.542 | 0.602 | 0.022 |
| 0.05 | 0.595 | 0.620 | 0.040 |
| 0.1 | 0.686 | 0.621 | 0.040 |
| 0.2 | 0.823 | 0.601 | 0.020 |
| 0.25 | 0.898 | 0.668 | 0.085 |
| 0.3 | 0.971 | 0.592 | 0.013 |
| 0.4 | 0.942 | 0.625 | 0.044 |
| 0.5 | 1.069 | 0.661 | 0.078 |
Standard curve: Y=1.0557X+0.5782 (R2=0.93); Take three of the highest concentration of UL128 purification liquid and condensed them into 0.1mg/mL and saved at −20°C for use.
Silver stain of recombinant UL128 protein. M: Marker, L1: Supernatant of CHO-Vector, L2: Purification lipid of CHO-Vector supernatant, L3: Blank control, L4: Supernatant of CHO-UL128; L5: Purification lipid of CHO-UL128 supernatant, rUL128: Western Blot of recombinant UL128 (using anti-Histag).
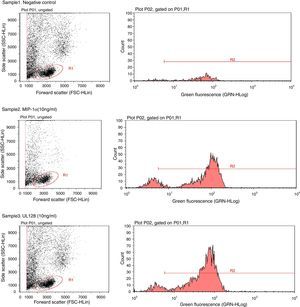
Chemotactic agonists induce a rapid and transient rise of Ca2+ in target cells. Our experiment showed that UL128 protein increased Ca2+ in PBMC at chemotactic concentrations which was similar to the function of MIP-1α (Fig. 2).
Sample1: The negative control group (PBMC+Fluo-3/AM). Fluorescent indicator did not detect significant Ca2+ when PBMC without treatment factor. Sample2: The positive control group (PBMC+Fluo-3/AM+MIP-1α). Intracellular Ca2+ concentration increased instantly when adding MIP-1α in the cell-culture medium. Sample3: The experimental group (PBMC+Fluo-3/AM+UL128). The level of intracellular Ca2+ concentration was markedly increased after treatment of UL128.
Immunohistochemistry results of cross-linking assay showed UL128 protein binding the cytomembrane receptors of PBMC. Human CC chemokine (MIP-1α) was used as positive control. The mean percentage of positive cells that UL128 and MIP-1αbinding with PBMC were 13% and 12.6%, respectively (Fig. 3). There was no significant difference between them (p>0.05).
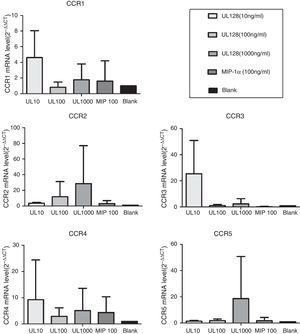
Expression of chemokine receptors in PBMCIn our experiment, UL128 protein could enhance production of CC chemokine receptors (CCR) according to PCR detection. CCR1, CCR3, and CCR4 were up-regulated when PBMC was co-cultured with UL128 at low concentration (10ng/mL). Transcription of CCR2 and CCR5 were improved with the increase of UL128 concentration, but they could hardly be detected with UL128 at low concentration. Based on these results, we infer that HCMV encoded UL128 protein may interact with different kinds of CCRs of PBMC and active at low concentration (Fig. 4).
DiscussionThe study further proved the immune modulatory function of a viral encoded CC chemokine (HCMV UL128). This viral encoded protein not only has similar structure, but also has similar function to the human CC chemokine MIP-1α, which is called chemokine mimics. In vitro, UL128 has the ability to attract PBMCs which is equal to that of MIP-1α and its chemotactic effect does not increase with the increasing of concentrations, which were shown in our previous study.7,8
Immune modulators regulate immunity through a series of complex processes with related polypeptides. Receptor binding is the first step of activation of immune cells. Chemokines exert their functions by interacting with seven-trans-membrane domain receptors expressed in different immune cell types, thus producing different types of inflammation.9 Here we demonstrated that HCMV encoded UL128 played a role in receptor binding and activation of PBMC. Calcium flux was observed in PBMC after addition of UL128 at chemotactic concentration (10ng/mL), which confirms that recombinant UL128 has the ability to activate PBMC in vitro. Moreover, both immunochemistry and PCR results demonstrated the capacity of UL128 to interact with the cell surface receptors of PBMC. Straschewski et al. reported that soluble UL128 protein (1000ng/mL) in vitro could block migration of monocytes by down-regulation of CCR1, CCR2 and CCR5 comparing with monocytes infected with wild type of HCMV.10 Nonetheless, in our study recombinant UL128 could enhance the production of CCR1 as well as CCR3 and CCR4 at low concentration (10ng/mL) which may correlate with its chemotactic function such as recruiting PMBC to the infectious site. It was predicted that only expression of chemokines at low levels in anatomically restricted areas could produce leukocytic infiltration consistent with their in vitro properties.4 The artificial high concentration of molecules in vitro may cause inhibitory effect by itself. Based on the above findings, we speculate that UL128 may combine with a variety of CC chemokine receptors and different concentrations of UL128 could enhance the expression of different receptors so as to having diverse influences to human immune cells. This may explain our previous findings why UL128 protein exerts chemotactic function without concentration dependence.
So far, five diverse functions of chemokine mimics have been identified: anti-chemokines; cell-entry factors; cell-growth factors; angiogenic factors and leukocyte chemoattractants.11 According to our studies, HCMV encoded UL128 acts as a chemokine agonist in vitro recruiting preferred host cells (leukocytes) to the site of infection and possibly to ensure viral latency at low concentration, and may cause imbalance of host immune system simultaneously. CC chemokine receptors 1, 3 and 4 may be involved in this process. The CC chemokine receptors have overlapping specificities, which makes the receptor–ligand research more confusing. Moreover, study of chemokines in vitro may not be enough to predict their in vivo properties. Recently, a genetic demonstration for the critical role of human β-chemokine MIP-1α in viral disease has been shown. Mice engineered to be deficient in MIP-1α show none of the presumably autoimmune myocarditis linked with Coxsackie virus infection. Similarly, pulmonary inflammatory responses to influenza virus were alleviated.12 They suggest that MIP-1α plays an irreplaceable role in the inflammatory responses to infection and clearance of virus. This may explain why HCMV encoded UL128 mimic human chemokine MIP-1α. Next step, we will move on to the detailed mechanism by which chemokine mimics and related pathology in the end organ damage of HCMV infection.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by National Science and Technology Support Program of China (grant no. 2012BAI04B05), National Natural Science Foundation of China (grant no. 81071337) and Natural Science Foundation of Zhejiang Province (grant no. Z2110006).