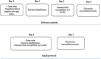
Antimicrobial treatment of patients with bloodstream infections (BSI) is time-sensitive. In an era of increasing antimicrobial resistance, rapid detection and identification of bacteria with antimicrobial susceptibility are critical for targeted therapy early in the disease course. This study describes the performance of a rapid method for identifying and testing antimicrobial susceptibility of Gram-negative bacteria performed directly from blood culture bottles in a routine microbiology laboratory. A total of 284, 120, and 24 samples were analyzed by rapid identification (Rid), rapid susceptibility testing (RAST), and rapid broth microdilution for polymyxin B (rMIC), respectively, and compared with standard methods. Our protocol was able to identify 93% of isolates at the species level using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). We obtained 100% agreement for RAST compared to the standard method and 96% agreement for rMIC. Our protocol has proven to be an excellent tool for rapid identification of Gram-negative bacilli causing BSIs. It can also be used in microbiology laboratory routine along with RAST and faster polymyxin microdilution, especially for carbapenemase-producing bacteria, allowing for rapid, simple, accurate, and cost-effective diagnosis.
Sepsis is a disruption of homeostasis in the human body in response to bloodstream infection (BSI) and is associated with a high risk of mortality.1 According to the World Health Organization (WHO), sepsis is one of the most common adverse events in healthcare settings, affecting approximately 30 million people worldwide and resulting in six million deaths.2 Timely detection of antimicrobial resistance factors is therefore crucial for the clinical management of BSI caused by Gram-negative bacteria. When BSI is suspected, the time between blood collection and identification of the microorganism, together with antimicrobial susceptibility results, represents a diagnostic window of uncertainty during which patients usually receive comprehensive empiric therapy.
Among BSIs, those caused by carbapenemase-producing enterobacterales (CPE) have the highest mortality rate within 30 days compared to those caused by other bacteria. Moreover, the number of cases of CPE infection in our institution has increased in recent years.3 Rapid identification of the causative agent may be critical for treatment efficacy. Studies suggest that administration of antibiotics in the first three hours after sepsis results in up to 14% lower mortality compared with patients who received antibiotics after this time period.4 However, conventional methods usually take at least 48 hours to report microbial identification and antimicrobial susceptibility after blood cultures become positive.5
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a technique for identifying microorganisms by detecting proteins.6 The use of MALDI-TOF MS in routine clinical microbiology laboratories contributes to faster identification of microorganisms compared to conventional methods and can lead to better and faster therapeutic interventions.7 Using this technology, the development of new, even faster methods for the identification of microorganisms has already been reported in the literature.8-10 However, these methods are laborious, expensive, and require chemical reagents that pose a risk to the environment and the health of laboratory workers. In addition to rapid identification of microorganisms, the combination of MALDI-TOF MS and a rapid antimicrobial susceptibility test (RAST) shows a more significant advantage than rapid bacterial identification alone.11 The aim of this study was to propose a new flowchart for processing positive blood cultures in routine clinical microbiology laboratories and to evaluate the performance of a rapid method for identification and antimicrobial susceptibility testing of Gram-negative bacteria directly from blood culture bottles.
Materials and methodsStudy design and populationThis study included positive blood cultures for aerobic bacteria from patients treated at Hospital de Clínicas de Porto Alegre between January and September 2020. Samples were incubated in an automated BacT/ALERT® 3D system (bioMérieux, France). Blood cultures with Gram-negative bacterial growth observed between 7 am and 2 pm were included in the study. Polymicrobial blood cultures were excluded, and only one series of blood cultures per patient was included in the analysis. In addition, only blood cultures from the microbiology laboratory were included: no artificially inoculated bottles were used in this study. The routine microbiology laboratory is located in an 840-bed university hospital (Hospital de Clínicas de Porto Alegre) in Porto Alegre, southern Brazil.
Standard identification method (sID) and standard antimicrobial sensitivity testing (sAST)Blood culture bottles were analyzed after the bacT/ALERT® 3D system flagged them as positive. Samples were analyzed by Gram staining followed by subculture on an appropriate solid agar medium (Chocolate agar or MacConkey agar, bioMérieux) after 18-24 hours of incubation at 35°C. Colonies grown on the plate overnight during incubation were mounted on a slide and prepared for analysis using VITEK MS® System software version 3.0 (bioMérieux) according to the manufacturer's instructions. The identification was considered correct if the confidence values were between 60 and 99.9, as indicated by the manufacturer. This system was referred to as sID. Colonies grown overnight on agar plates were also used to prepare an inoculum for the disk diffusion test (Kirby-Bauer disk diffusion test protocol) according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST), version 10. This disk diffusion method was referred to as sAST. Bacterial identification and antimicrobial resistance results obtained using this conventional workflow were used for comparison in data analysis. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control.
Polymyxin B minimum inhibitory concentration (MIC) — the standard method (sMIC)Susceptibility to polymyxin B (test range 64-0.125 μg/mL) was tested by broth microdilution in cation-adjusted Mueller-Hinton broth (CA-MHB, Beckton Dickinson, USA). Results were evaluated after 18-24 hours and interpreted according to the clinical breakpoints of EUCAST, version 10.0. Isolates with a MIC of polymyxin B ≤ 2 μg/mL were considered sensitive and those with a MIC of > 2 μg/mL were considered resistant. E. coli ATCC 25922 and P. aeruginosa ATCC27853 were used as quality control.
Rapid identification method (rID) and rapid antimicrobial sensitivity testing (RAST)rIDFor bacterial identification, the test was performed as follows: 3 mL of the positive blood culture broth was removed from the blood culture bottles with a sterile syringe and transferred to a 10-mL serum separator tube. The contents were centrifuged at 3000 rpm for five minutes, the supernatant was discarded, 3 mL of saline was added, and the sample was centrifuged at 3000 rpm for another five minutes. The supernatant was discarded and 1 μl of the pellet was added in duplicate to the slide spot. After drying at room temperature, 1 μL of matrix was added. Bacterial identification was then performed using the VITEK MS® system.
RASTThis method is currently validated for Escherichia coli, Klebsiella pneumoniae, P aeruginosa, and Acinetobacter baumannii complex by EUCAST RAST method version 2.1, dated April 2020. This assay was performed with positive blood cultures for these bacteria according to the method EUCAST RAST. Briefly, approximately 125 µL of blood culture was seeded directly from the bottle onto a Mueller-Hinton 90-mm agar plate (bioMérieux), an antimicrobial meropenem disk (10 µg) was added, and the plate was incubated at 35° C for 4-6 hours. Evaluation was performed according to the method EUCAST RAST, as follows: E. coli plates with meropenem zone diameters < 15 mm and K. pneumoniae plates with meropenem zone diameters < 13 mm after four hours of incubation were considered resistant. P. aeruginosa or A. baumannii isolates were considered resistant if the meropenem inhibition zones were < 14 mm and < 15 mm, respectively, after six hours of incubation.
Rapid polymyxin B MIC (rMIC)Susceptibility testing by broth microdilution was performed according to the EUCAST description and with some modifications. Meropenem-resistant isolates identified by RAST were selected for the rapid broth microdilution method. Inoculum was prepared from the rapidly grown culture (4-6 hours agar plate incubation) on Mueller-Hinton agar (bioMérieux), and plates were incubated overnight at 35°C ± 1°C. MIC was analyzed and results were interpreted according to EUCAST breakpoints12
The comparison between turnaround times of the standard and the fast protocols is shown in Fig. 1.
Data analysisBacterial identification and antimicrobial resistance results obtained with the modified rapid methods were compared with those obtained with the standard methods. Bacterial identification results were classified as correct at the species or genus level, with calculation of non-reliable identification and success rates.
Sensitive, intermediate, and resistant interpretation results were evaluated for each antimicrobial agent tested by both the disk diffusion and broth microdilution methods; categorical agreement (CA) and agreement (EA) were determined. EA Values (results within ± 1 doubling dilution of MIC) were determined using the reference method according to ISO 20776-1. Categorical discrepancies were classified as follows: very major errors (VME) or false-susceptible results; major errors (ME) or false-resistant results; and minor errors (mE) when one method gave an intermediate result and the other gave a susceptible or resistant result. The acceptable categorical discrepancy rates between the VME, ME, and mE methods were ≤ 1.5%, ≤ 3%, and ≤ 10%, respectively, according to the Food and Drug Administration (FDA).
ResultsAfter excluding 20 polymicrobial blood cultures, 284 samples were analyzed for rapid bacterial identification, and 19 different bacterial species were detected. The overall concordance rate of the rID method at the species level was 93% (264/284 samples). The highest concordance rate was observed for Enterobacterales species (92%), K. pneumoniae and E. coli (97% and 100% concordance, respectively). P. aeruginosa and the A. baumannii complex had 95% and 86% concordance in bacterial identification, respectively. Eighteen (6.33%) isolates could not be identified by the rID method (no organism detected), and only two isolates (0.7%) were misidentified: an Enterobacter cloacae complex was identified as Serratia marcescens, and a Salmonella group was identified as Cronobacter malonaticus (Table 1).
Comparison between rapid and reference identification methods.
For the RAST method, 120 samples were analyzed: 48 E. coli, 47 K. pneumoniae, 18 P. aeruginosa, and 7 A. baumannii complex. All RAST results were consistent with the standard protocol (sAST), and all 120 isolates had 100% of CA when comparing RAST and sAST protocols. Therefore, no VME, ME, or mE were observed (Table 2).
All isolates resistant to meropenem (n = 24) were tested for polymyxin B by the rMIC method: 16 K. pneumoniae, 3 P. aeruginosa, and 5 A. baumannii complex. For K. pneumoniae, rMIC50/rMIC90 values were 0.5/16 mg/L. For P. aeruginosa, rMIC50/rMIC90 values were 0.5/0.5 mg/L; for the A. baumannii complex, rMIC50/rMIC90 values were 0.5/0.5 mg/L. For all species, rMIC50/rMIC90 values were 0.5/16 mg/L. EA and CA were 96%, and no VME or mE were observed. One ME (0.24%) occurred with a K. pneumoniae isolate in which the standard method determined MIC = 2 (sensitive) and our technique determined MIC = 4 (resistant). The results are shown in Figs. 2-5.
DiscussionDuring our 9-month study at Hospital de Clínicas de Porto Alegre, an 840-bed tertiary care hospital in southern Brazil, 230 intensive care unit beds, we validated a 15-minute protocol that identified 93% of Gram-negative bacteria found in blood cultures to the species level. Our protocol was associated with RAST for meropenem on day 0 and provided polymyxin B MIC results on day 1, reducing turnaround time to approximately two days.
Our study was conducted over a long period of time and included many samples from patients in routine care. We replaced the use of chemical reagents, common to most techniques,10,13-15 with saline, a solution available in any microbiology laboratory that does not harm the environment or laboratory personnel. The technique was inexpensive, easy to perform, and identification results could be obtained within 15 minutes after positive labeling of the blood culture. It is important to highlight that our study was performed using a MALDI TOF VITEK® MS from bioMérieux, in contrast to most published studies performed using Bruker Daltonics' MALDI TOF MS.10,13,16,17 MALDI-TOF MS is an efficient and cost-effective method for bacterial identification compared to routine biochemical panels.18 Its use in rapid identification of pathogens causing sepsis is essential to provide clinicians with information to decide on appropriate therapy.19 Direct identification from blood cultures can reduce the time to results to about 24 hours compared with identification from subcultured colonies.20 However, blood cells can interfere with the detection of bacterial proteins and usually need to be removed with lysis buffers.21
Because the identification technique was performed in parallel with routine procedures, we tested the spots once in duplicates. This could explain why some organisms were not detected: If the spot was not suitable for MALDI-TOF MS analysis, we did not repeat the test with the same sample. Had the samples been repeated, we could have obtained even better results in identifying bacteria. In addition, our results were similar to those of other studies that performed rapid identification directly from blood culture bottles containing Gram-negative bacilli;10,16,17,22 this indicates that our protocol is a viable technique for reducing the turnaround time of blood cultures.
This study showed 100% agreement when comparing RAST with the standard antimicrobial susceptibility test (AST) in 120 samples. We have shown that for bloodstream infections caused by E. coli, K. pneumoniae, P. aeruginosa, and the A. baumannii complex, it is no longer necessary to wait 24 hours for colony growth and more than 24 hours for AST results, as is the case in most clinical microbiology laboratories, because numerous studies confirm the reliability of RAST.23 In addition, the RAST is equally easy to perform and requires no more input than that already used for the standard AST.
The Global Action Plan on Antimicrobial Resistance developed by the World Health Organization24 includes the development of new diagnostic tools for antimicrobial susceptibility as one of its goals to reduce bacterial resistance. A study conducted by Baltas et al.25 in 2020 showed a survival benefit for patients who received effective antibiotic treatment early compared with those who did not; the latter ultimately had a higher risk of death. This study also showed that the most common reason for ineffective treatment was antimicrobial resistance, reinforcing the need for faster ASTs.
Finally, we used short incubation colonies to perform broth microdilution for polymyxin B on the day that blood cultures were labeled positive. This allowed us to reach 97% of EA and obtain the final results in less than 24 hours. It is important to note that the only isolate that changed sensitivity category had an MIC = 2, which is at the borderline between sensitive and resistant strains; taking into account the ± 1 dilution, the result for this isolate was consistent with the standard method.
By incorporating these three techniques into the laboratory routine, we were able to identify the bacteria and determine their susceptibility profiles in approximately four to six hours after positivity. If the microorganism was resistant to meropenem, we were able to inform clinicians of the MIC of polymyxin B and initiate the most appropriate antimicrobial therapy in less than 24 hours after positivity.
We previously have proposed a faster, easier, and reliable method to reduce the turnaround time of bloodstream infection diagnosis in a routine microbiology laboratory.25 The difference between the proposed method and the standard ones was centrifugation and short-time growth on solid media. However, timely pathogen detection and availability of susceptibility data are essential for optimal treatment, thus in this study we upgraded the method of direct identification and antimicrobial susceptibility testing of Gram-negative bacteria from positive blood cultures. This eliminates the need for initial broth subculture and incubation to obtain isolated colonies for further testing.
To the best of our knowledge, this is the first study to combine three different techniques: rID, RAST, and polymyxin B rMIC. Our results show that this protocol can be used in a routine clinical microbiology laboratory and can replace standard protocols, saving at least 40 hours for the final result compared with reference methods.
Limitations of our study include the small number of carbapenem-resistant bacteria available to perform the broth microdilution technique and the lack of an evaluation of the impact of this technique on patient recovery. However, other studies in this context14,26 have shown that our protocol is very promising in that regard.
In conclusion, our protocol is an excellent tool for the rapid identification of Gram-negative bacilli causing bloodstream infections using MALDI-TOF MS. It can also be used in a clinical microbiology laboratory routine with a rapid sensitivity test and faster polymyxin B microdilution to obtain a rapid, simple, accurate, and inexpensive diagnosis. It is worth noting that after this study, we have implemented this workflow into our laboratory routine and we will soon be able to evaluate the impact of this change in our facility.
FundingThis work was supported by the Research Incentive Fund [FIPE/HCPA], Porto Alegre - Brazil.
Ethical approvalApproval was obtained from the ethics committee of Hospital de Clínicas de Porto Alegre (CAAE:23747219000005327).
The authors would like to thank the Research Incentive Fund [FIPE/HCPA].