Colonizations/Infections caused by carbapenem-resistant Enterobacterales are of great clinical and epidemiological importance due to their rapid dissemination and high mortality rates. In this scenario, the use of antibiotics intensified by the COVID-19 pandemic has brought about a great warning on the real impact that this pandemic could have on antimicrobial management programs and long-term antimicrobial resistance rates. The objective of this study was to evaluate the increase of New Delhi Metallo β-Lactamase (NDM)-producing Enterobacterales cases in COVID-19 units of a complex Brazilian tertiary hospital. This retrospective observational study included all patients admitted to the hospital identified as colonized or infected by NDM-producing Gram negative bacilli (GNB), from January 2017 to April 2021. Forty-two NDM-producing Enterobacterales were identified in 39 patients. The rate of NDM cases per total surveillance cultures increased progressively between 2017 and 2021 (chi-2 for trend, p < 0.0001) and was associated with a higher occurrence specifically in COVID units (Fisher exact, p < 0.0001). The molecular investigation of the NDM-producing Klebsiella pneumoniae strains revealed the emergence of diverse clones during the COVID-19 period, also with possible evidence of horizontal transmission among patients within COVID units. NDM-producing Enterobacterales with multiple and different clonalities in the COVID-19 units also raised questions about the importance of other factors besides horizontal clonal transfer, including the increase of antimicrobial consumption by these patients.
New Delhi Metallo β-lactamases (NDM) production has been progressively identified in carbapenem-resistant Gram-negative bacteria on all continents, representing a significant challenge for clinical management and public health worldwide.1,2 Since 2013, the increase of NDM-positive strains in Brazil has been described in different species of Gram-negative bacilli (GNB) recovered from human, animal, and environmental sources.2-5 The transmission of NDM-positive strains in health care settings is related to close contact, with hospitalized patients being the most vulnerable for colonization and infection. Although, in general the same infection control measures should be applied to any carbapenem-resistant organisms regardless of the resistance mechanisms,6 NDM- and Klebsiella pneumoniae carbapenemase (KPC)-producing isolates apparently present relevant epidemiological differences.1,2 While KPC-producing isolates have been widely described internationally belonging to a major well-adapted K. pneumoniae clonal complex (258),7 NDM-producing isolates are commonly described in multiple GNB8 with apparently no sustained global spread of specific high-risk clades.1
Recently, the increased use of antibiotics may have been intensified by the COVID-19 pandemic worldwide9,10 raising concerns about the real impact of the pandemic on antimicrobial management programs and long-term antimicrobial resistance (AMR) rates,9 particularly strategies directed to COVID-19. The impact of the COVID-19 pandemic in Brazil and in São Paulo has posed significant threats to all, and in particular to the public hospital system10 in terms of allocated resources and quality of care. Most public hospitals have dedicated specific units (wards and intensive care units - ICU) for accommodating suspected and confirmed COVID-19 patients. In the study institution, the first case of carbapenemase detection (KPC) occurred in 2009 and spread in the postoperative ICU followed by some outbreaks of an endemic cluster of Klebsiella pneumoniae clonal complex 258 in the hospital environment.11,12 In this report, we describe the increase of another type of carbapenemase designated as NDM-1 producing Enterobacterales in COVID-19 units of a Brazilian high complexity tertiary hospital.
Material and methodsPopulation samplesThis was a retrospective observational study based on surveillance cultures and target medical units conducted at Dante Pazzanese Institute of Cardiology (IDPC), a tertiary 350-bed hospital specialized in cardiovascular surgery. IDPC was included in the public COVID-19 efforts established in March 2020. As part of the efforts to manage the pandemic, IDPC infection control service and hospital administration established specific units and flows devoted to COVID-19 patients, while maintaining other units for their regular cardiothoracic patients. In the present study, we categorized these COVID-19 units (ICU or specific wards) and General units (ICU and Wards for non-COVID-19 patients). Since 2013, the infection control team performs weekly routine inguinal and anal surveillance cultures at the ICU, emergency room, and wards searching for carbapenem-resistant Enterobacterales (CRE), carbapenem-resistant non-glucose-fermentative Gram-negative bacilli (BGN-NF) (Acinetobacter baumannii and Pseudomonas aeruginosa) and vancomycin-resistant Enterococci (VRE), with the objective of placing those colonized patients in contact precautions. All patients admitted to COVID and non-COVID-19 units from January 2017 to April 2021, and identified as colonized or infected by NDM-producing GNB, were included in this study.
Microbiological methodsAll surveillance cultures growing GNB were submitted to bacterial identification by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF/MS) technology, using Vitek MS® system (bioMeriéux, Marcy-I’Étoile, France). The minimum inhibitory concentration (MIC) of all antibiotics was determined using Vitek®2 System (bioMeriéux), except for polymyxin B which was done by broth microdilution (Probac, Brazil). All surveillance cultures with GNB identified during the study period were screened for carbapenemase presence using commercially available disks containing carbapenems with and without EDTA (0.1 M), cloxacillin (75 mg/mL) or phenylboronic acid (40 mg/mL), as recommended by the Brazilian National Health Surveillance Agency. If screened positive, a sample was submitted to confirmation by Real-Time PCR for the detection of the following carbapenemase genes: blaKPC,blaNDM, blaIMP, blaVIM, blaGES, and blaOXA-48-like.13
Strains of the most prevalent NDM-producing species were analysed by Pulsed-Field Gel Electrophoresis (PFGE) for genetic relatedness, using the restriction endonuclease SpeI (New England Biolabs Inc., Ipswich, MA). PFGE profiles were analyzed and compared using BioNumerics version 8 software (Applied Maths, Sint-Martens-Latem, Belgium) and cluster analysis was performed with the unweighted pair group method with arithmetic mean, Dice coefficient and pattern interpretation. Isolates that exhibited a PFGE profile with ≥ 80% similarity were considered closely related.
Statistical analysisA single NDM-producing isolate per patient was considered for the descriptive and statistical analysis. Information on variables age and sex of patients, NDM identification unit type (COVID or non-COVID-19), patient length-of-stay until NDM detection, surveillance culture, colonization or infection by NDM-producing GNB, and outcome (discharge or death) was collected. GraphPad Prism version 7.05 (San Diego, CA) was used for the chi-squared test for trend on the rate of new NDM patient (colonization's or infections) per the total number of surveillance cultures per year, from 2017 to 2021, and the Fisher's exact test on the rate of new NDM patient (colonization's or infections) per the total number of surveillance cultures per unit, COVID or non-COVID-19.
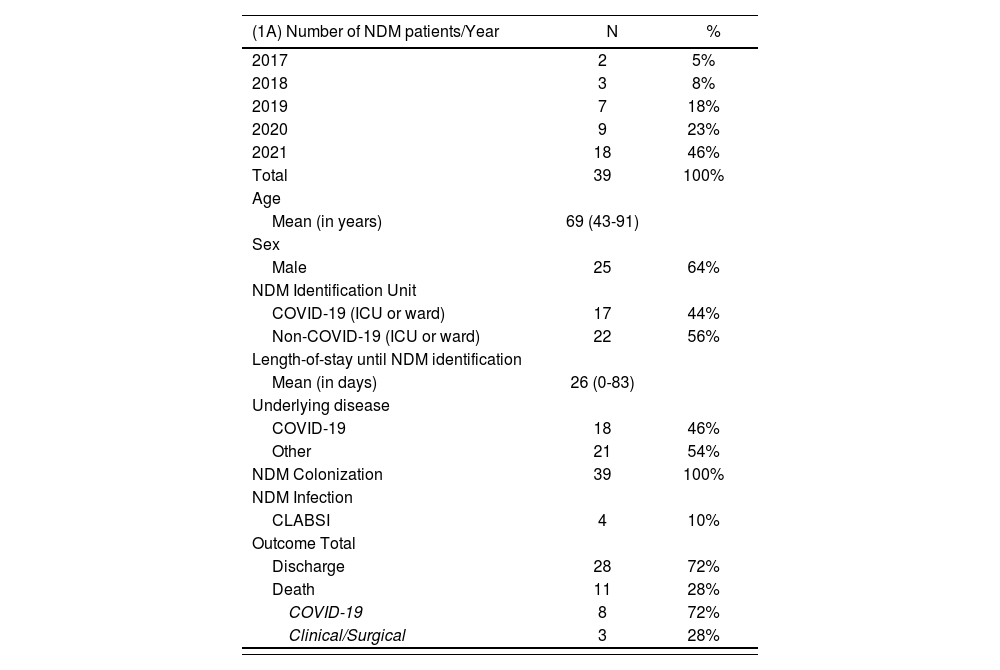
ResultsA total of 13,004 active surveillance cultures were performed between January 2017 to April 2021. Nine hundred twenty-nine (7.1%) were positive for multidrug-resistant (MDR) bacteria: 616 (66%) CRE, 176 (28.5%) VRE, and 137 (22%) BGN-NF. Among CRE samples, there were 42 (6.9%) NDM-producing Enterobacterales isolated in 39 patients: 36 Klebsiella pneumoniae, two Enterobacter cloacae, two Proteus mirabilis, one Morganella morganii and one Providencia rettgeri. Three patients carried more than one NDM-producing in different species of Enterobacterales. From the 39 patients identified with at least one NDM-producing GNB during the study period, all were considered colonized (rectal or inguinal sites) during the hospital stay and 4/39 patients presented central line-associated bloodstream infection (CLABSI). The time between colonization and infection ranged from four to seven days. Table 1A shows patient characteristics.
(A) Total New Delhi Metallo β-Lactamase (NDM)-producing GNB cases detected per year and patient characteristics, and outcome (discharge or death). (B) Distribution of 36 NDM-producing Klebsiella pneumoniae strains identified in the present study according to their PFGE pattern, hospital unit and year of detection.
All 42 NDM-producing Enterobacterales had no remaining carbapenemase genes evaluated and showed high-level of resistance to broad-spectrum cephalosporins, carbapenems, ciprofloxacin, gentamicin and piperacillin-tazobactam, according to CLSI and BrCAST guidelines.14,15 All K. pneumoniae samples (n = 36) were susceptible to polymyxin B (MIC ≤ 0.5 µg/mL) according to BrCAST or EUCAST guidelines.15,16
Table 1B shows the PFGE method performed only on the Klebsiella pneumoniae isolates (n = 36) (one sample per patient). The dendrogram analysis revealed 17 PFGE patterns (“A” to “Q”). Five clusters (F, H, J, L and N) were detected with more than 80% of similarity by the Dice coefficient (Table 1B). The most prevalent cluster “N” was detected in 12 patients with 87.6% of similarity. Three of 12 K. pneumoniae strains cluster “N” was detected before the COVID pandemic period (year 2019), and nine strains were detected during the COVID pandemic period (after March 2020) and within COVID units. Cluster “H” was found in three patients in COVID units at the same period with 100% similarity. Cluster “L” was detected in three patients in non-COVID units.
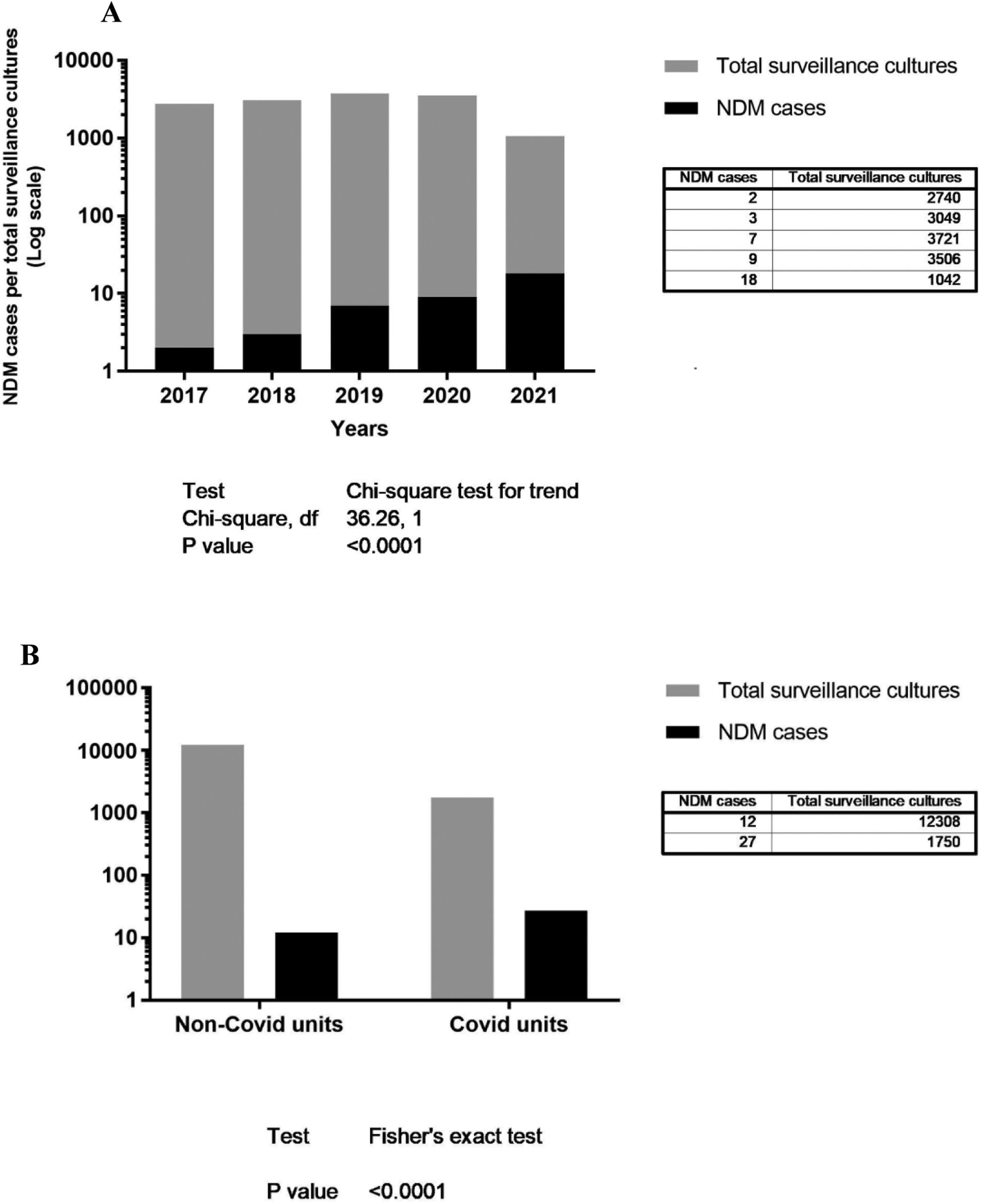
Fig. 1A shows the rate of NDM cases (colonizations or infections) per total surveillance cultures from 2017 to 2021, with the respective total amounts and chi-squared test for trend (p < 0.0001), and 1B the rate of NDM cases (colonizations or infections) per total surveillance cultures per COVID or Non-COVID units, with the respective total amounts and Fisher's exact test (p < 0.0001).
Discussion and conclusionThe COVID-19 pandemic posed a significant threat, not yet overcome, on health systems worldwide, and despite relevant public responses, public hospitals have faced important secondary impacts on their structure, particularly on increased density of inpatients, extenuated health care workers, and increased antimicrobial consumption.10,17,18 In the pandemic scenario (March 2020 to December 2021), to enable hospitalization of COVID-19 cases, the IDPC reduced the number of elective cardiovascular surgeries, setting up 30 ICU beds and 40 designated beds for COVID-19 patients. There was a 20% reduction in hospitalizations compared to previous years. Elective surgeries were suspended during part of this period, hence leading to a 30% reduction in the number of procedures. However, even with the reduction of hospitalizations, there was still an increase in the consumption of antimicrobials, according to hospital pharmacy information. In the period of January to December 2020, ceftriaxone consumption increased by 22%, piperacillin-tazobactam by 14%, and azithromycin by 70%. In a cohort of 52 critically ill patients with SARS CoV-2 pneumonia in hospital care in Wuhan city, the rate of broad-spectrum antibiotics use was 94%, although the reported incidence of secondary infections was lower than 15%.19
In the present study, we report five years of surveillance to multidrug resistant GNB in a complex hospital, with a significant increase in NDM-producing bacteria over time. There was a significant increase detected by chi-squared test for trend on NDM-producing GNB (p < 0.0001) between 2017 and April 2021, specially in COVID-19 units (p < 0.0001) (Fig. 1). All isolates were highly resistant to the antimicrobial agents tested, and unfortunately, polymyxin B was considered the last resort to treat these patients. The emergence of such antimicrobial resistance pattern (including readily transmissible mobile genetic elements) in most of the cases is caused by suboptimal and/or excessive use of antimicrobials inside and outside hospital settings. Suboptimal antimicrobial usage often stems from the absence of or inappropriate interpretation of microbiological diagnosis.20
The PFGE analysis showed the emergence of different clusters after March 2020, which coincides with the beginning of COVID-19 pandemic. Different clusters were identified during these periods, one of them (Cluster H) exclusively in COVID-19 units and another (Cluster L) exclusively in non-COVID-19 units (Table 1B). Among 17 patients admitted in COVID-19 units, nine were colonized by the same NDM-producing K. pneumoniae cluster “N” and three were colonized by cluster H. These findings are reasonable evidence of horizontal NDM-producing K. pneumoniae transfer occurring between patients admitted in COVID-19 units, which may be attributed to the pandemic pressures of higher patient density, new and young healthcare employees at COVID-19 units, work overload even though the infection control team have performed several trainings to all professionals, including HAI infection prevention and isolation precautions.
The diversity of NDM-producing Enterobacterales species and K. pneumoniae clones observed in the present study brings up an interesting question. As previously shown, NDM-producing isolates are commonly described in multiple GNB8 with apparently no sustained spread of specific high-risk clades.1 In addition, blaNDM genes have also been detected in hospital sewage of several countries, which may reflect intestinal carriage of NDM-producing strains among the population of health care settings.1 Thus, it seems reasonable to hypothesize that the cluster diversity and the major differences in clones detected pre- and post-March 2020 may have been associated to other factors, not only related to patient density and horizontal transmission. In this scenario, antimicrobial consumption might have provided the necessary selective pressure for the emergence of colonization by multidrug-resistant pathogens carrying extended-spectrum beta-lactamase genes, including carbapenemase genes. Further studies should be carried out to clarify these findings.
FundingThis work was supported by AFIP – Associação Fundo de Incentivo a Pesquisa.
Ethical approvalThe study was approved by the Ethical Committee of Instituto Dante Pazzanese de Cardiologia (São Paulo, Brazil) [n° 5162].
We would like to thank Henri Berghs for kindly providing us the BioNumerics Software used in this study, Vera Lucia Barbosa and Paulo H. D. Santos for the collection of epidemiological data, and Ana Paula lobo for support in the laboratory.