The mechanism underlying the coexistence of hepatitis B surface antigen and antibodies to HBsAg in chronic hepatitis B patients remains unknown.
AimsThis research aimed to determine the clinical and virological features of the rare pattern.
MethodsA total of 32 chronic hepatitis B patients infected by HBV genotype C were included: 15 carrying both HBsAg and anti-HBs (group I) and 17 solely positive for HBsAg (group II). S gene and reverse transcriptase region sequences were amplified, sequenced and compared with the reference sequences.
ResultsThe amino acid variability within major hydrophilic region, especially the “a” determinant region, and within reverse transcriptase for regions overlapping the major hydrophilic region in group I is significantly higher than those in group II. Mutation sI126S/T within the “a” determinant was the most frequent change, and only patients from group I had the sQ129R, sG130N, sF134I, sG145R amino acid changes, which are known to alter immunogenicity.
ConclusionsIn chronic patients, the concurrent HBsAg/anti-HBs serological profile is associated with an increased aa variability in several key areas of HBV genome. Additional research on these genetic mutants are needed to clarify their biological significance for viral persistence.
Hepatitis B virus (HBV) infection is a serious global public health problem, and its prevalence varies considerably from region to region. Worldwide, more than 350 million people are chronic carriers of HBV, and have a risk of dying from development of cirrhosis and hepatocellular carcinoma.1 Clinically, the appearance of circulating hepatitis B surface antigen (HBsAg) heralds HBV infection, whereas the presence of the antibody to HBsAg (anti-HBs) usually indicates resolution of infection and is considered indicative of immunity to HBV infection.2 It is generally believed that anti-HBs can neutralize and clear HBsAg. Therefore, the concomitant presence in the same serum of HBsAg and anti-HBs could be possible. However, the persistence of HBsAg associated with anti-HBs in patients with chronic HBV infection has been reported in previous studies.3–5 So far, the mechanism underlying the concurrent detection of HBsAg and anti-HBs remains largely controversial.
HBV genome contains four partially overlapping open reading frames (ORF), and the “a” determinant is located at codon positions 124–147 within the major hydrophilic region (MHR) of the HBsAg. This determinant is the main target of recognition of HBsAg by anti-HBs and immune response cells during the course of the initial immune response in acute hepatitis B. HBV has been classified into eight genotypes, designated as A–H, and the most prevalent genotypes in China are genotypes B and C.5 Notably, previous reports documented that, in comparison to genotype B, genotype C takes a more aggressive disease course and has a lower response rate to antiviral therapy.6–9
Several reports showed the paradoxical coexistence of HBsAg and anti-HBs might be associated with the selection of HBV immune escape mutants during chronic carriage.10–12 These studies documented increased amino acid (aa) mutations in and around the “a” determinant, which is responsible for the stability and immunogenicity of HBsAg.13 Nonetheless, a later report rejected this hypothesis and instead suggested that the pattern of simultaneous appearance of HBsAg and anti-HBs was related to the weak binding of anti-HBs to HBsAg.12 As the reverse transcriptase (RT) region of HBV polymerase gene overlaps the HBsAg ORF, mutations within HBsAg gene might result in structural and functional alterations in the HBV reverse transcriptase with potential influence on viral replication capacity and efficacy of antiviral drugs.14,15
Therefore, the aims of this study were to investigate the prevalence of the coexistence of anti-HBs in HBsAg-positive CHB patients infected with genotype C HBV, and explore the relationship between the variability of the HBV S gene and the paradoxical serological profile.
Materials and methodsPatientsFrom January to December 2013, 1194 patients with CHB recruited from Wuhan, Huangshi, and Yichang were enrolled in the study. The inclusion criteria were HBsAg carriage for more than six months, HBV DNA concentration of >1×104IU/mL, serum alanine aminotransferase (ALT) >2 ULN (upper limit of normal), and associated symptoms. Exclusion criteria included having received passive immunization, previous antiviral therapy, coinfection with other hepatitis viruses, or HIV. Sera were collected and stored at −70°C. All individuals provided written informed consent before entering the study.
Serologic testingHBV serological markers were determined by using the chemiluminescent microparticle immunoassay (CMIA) technique with an Architect-i2000 automatic analyzer (Abbott Laboratories, USA). Commercially available kits were purchased from Abbott Laboratories. The analytical threshold of anti-HBs was defined as 10mIU/mL. Serum was diluted according to the Manual Dilution Procedure if HBsAg concentration value >250,00IU/mL. ALT was assayed with an ADVIA automatic biochemical analyzer (Siemens, Germany).
HBV DNA quantificationHBV DNA was quantified in serum using a commercially available real time fluorescence quantitative kit (Da An, China) with a lower detection threshold of 500copies/mL.
Serum DNA extractionViral DNA was extracted from 200μL of serum using QIAamp DNA blood mini kit (Qiagen, Germany) according to the manufacturer's instructions.
PCR amplification and sequencing of PCR fragmentAmplification and sequencing of the full-length S gene and the overlapping RT regions were performed using the protocol as follows. The primer sequences were synthesized by Beijing SBS Biotechnology Company. PCR was carried out in a 50-μL reaction mixture containing 10μL of HBV DNA template, 0.2μM each primer, 0.2mM of each dNTP, 2mM MgCl2, and 1μL of high-fidelity Taq polymerase (Takara, China). Besides, PCR was performed with hot start and denaturation at 94°C for 5min, 35 cycles at 94°C for 1min, at 50°C for 1min, and at 72°C for 2min, then incubated at 72°C for 5min. In order to obtain a complete description of HBV quasispecies, sequence analysis was performed with the powerful ultra-deep sequencing (UDS) approach, using the primers according to the reference sequence X04615 (the upstream primer: 5′-GTCACCATATTCTTGGGAAC-3′ nt2818-2837; the downstream primer: 5′-CATATCCCATGAAGTTAAGG-3′ nt 888–869).12
Sequence analysisGenomic sequences obtained for the S gene and the overlapping RT regions were translated into aa sequences and compared with HBV reference sequences used on the NCBI Website (http://www.ncbi.nih.gov/projects/genotyping/view.cgi?db=2). For analysis, the full-length S protein was divided into three regions corresponding to structural and/or functional domains: the N terminal region (aa 1–99), the MHR (aa 100–169), which contains the “a” determinant (aa 124–147), and the C terminal region (aa 170–226).
Statistical analysisData were stored using Microsoft Excel and analyzed using SPSS 17.0 software (Chicago, IL). Quantitative data were compared using Mann–Whitney or t test, and mutation rates were analyzed with the Chi-square test. Statistical significance was determined at a p-value <0.05.
ResultsCharacteristics of patientsOut of the 1194 CHB patients enrolled in this study, in 36 (3.0%) HBsAg and anti-HBs were concomitantly detected. In order to elucidate the mechanisms of coexistence of HBsAg and anti-HBs in patients infected with HBV of the genotype C, two groups were set up on the basis of their serological profile.
Fifteen HBsAg+/anti-HBs+ CHB patients infected with genotype C HBV were enrolled as the experimental group (group I). Meanwhile, 17 HBsAg+/anti-HBs− CHB patients infected with genotype C HBV were selected as the control group (group II). None of the patients had neither received vaccination against HBV nor antiviral therapy.
Table 1 shows the clinical characteristics of group I. There were no significant differences between group I and group II with regard to age, gender, serum ALT level, HBsAg level, HBeAg positivity rate, HBV DNA concentration (p=0.127, 0.755, 0.692, 0.610, 0.388, 0.234, respectively) (Table 2). Importantly, the patients with anti-HBs had rather higher anti-HBs level, with a median titer of 36.9mIU/mL (range, 18.52–1326.5).
Summary of clinical features of patients with concurrent HBsAg and anti-HBs.
| Patient | Gender/age | HBsAg (IU/mL) | Anti-HBs (mIU/mL) | ALT (IU/L) | HBeAg | HBV-DNA |
|---|---|---|---|---|---|---|
| P1 | F/23 | 325.6 | 1326.5 | 224 | Pos | 8.63E+07 |
| P2 | M/40 | 103.19 | 51.21 | 95 | Neg | 5.35E+05 |
| P3 | M/27 | 102.28 | 156.8 | 168 | Pos | 5.36E+06 |
| P4 | F/21 | 179.88 | 36.9 | 147 | Pos | 5.20E+06 |
| P5 | M/30 | 167.17 | 23.6 | 103 | Neg | 2.36E+04 |
| P6 | M/29 | 122.47 | 111.25 | 115 | Pos | 3.12E+06 |
| P7 | F/31 | 85.72 | 62.36 | 98 | Pos | 6.45E+06 |
| P8 | F/22 | 669.81 | 20.65 | 214 | Pos | 2.80E+08 |
| P9 | M/29 | 92.28 | 18.52 | 136 | Pos | 6.35E+06 |
| P10 | M/31 | 116.6 | 26.32 | 94 | Neg | 3.68E+05 |
| P11 | F/25 | 522.53 | 19.62 | 197 | Pos | 5.61E+07 |
| P12 | M/35 | 93.18 | 85.26 | 115 | Pos | 3.26E+05 |
| P13 | M/28 | 72.01 | 35.15 | 94 | Pos | 3.72E+04 |
| P14 | F/31 | 126.07 | 25.36 | 97 | Neg | 4.35E+05 |
| P15 | F/33 | 718.63 | 120.56 | 319 | Pos | 3.16E+09 |
M, male; F, female; Pos, positive; Neg, negative; ALT, alanine aminotransferase.
Comparative analysis of clinical features of patients between group I and group II.
| Group I(n=15) | Group II(n=17) | p | |
|---|---|---|---|
| Age, median years (range) | 29 (21–40) | 30 (18–43) | 0.127 |
| Gender, female:male | 7:8 | 7:10 | 0.755 |
| HBsAg titer, median IU/mL (range) | 122.47 (72.01–718.63) | 119.37 (68.25–625.53) | 0.610 |
| HBeAg+ (%) | 73.3 | 58.8 | 0.388 |
| Serum ALT level, median IU/L (range) | 115 (94–319) | 147 (92–325) | 0.692 |
| Serum HBV DNA level, median (log10copies/mL) | 6.71 (4.37–9.50) | 5.57 (4.42–8.71) | 0.234 |
ALT, alanine aminotransferase.
The entire S encoding gene sequences for patients with and without anti-HBs were available by PCR amplification and sequencing, and subsequently compared to reference sequences of the same genotype (Figs. 1 and 2 and Table 3). Thirteen patients from group I had single or multiple aa substitutions within the S protein.
Amino acid substitutions within HBsAg and the overlapping regions of reverse transcriptase from patients with concurrent HBsAg and anti-HBs. The part within the “a” determinant is underlined. Reference sequences of genotype C are obtained from the NCBI website (http://www.ncbi.nlm.nih.gov/projects/genotyping/genotype C: X046151). A short line demonstrates there is no mutation in this locus, and “stop” represents the stop codon.
Number of substitutions per 100 amino acid positions within HBsAg and overlapping regions of reverse transcriptase.
| Region of HBsAg | HBsAg | HBsAg overlapping regions of reverse transcriptase | ||||
|---|---|---|---|---|---|---|
| Group I | Group II | p | Group I | Group II | p | |
| Complete HBsAg | 1.45 | 0.55 | 0.000 | |||
| Complete RT | 0.71 | 0.26 | 0.005 | |||
| N-terminal Region | 1.01 | 0.42 | 0.044 | 0.34 | 0.18 | 0.595 |
| MHR | 2.67 | 0.67 | 0.000 | 1.14 | 0.17 | 0.003 |
| a-Determinant | 4.12 | 1.47 | 0.022 | 2.22 | 0.49 | 0.073 |
| C-terminal Region | 0.70 | 0.62 | 0.828 | 0.82 | 0.52 | 0.425 |
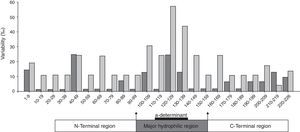
The distributions of aa substitutions were heterogeneous along the S protein (Fig. 2). Apparently, the highest frequency of aa substitutions was observed within the “a” determinant for both groups. Besides, aa variability within the MHR was markedly higher than that in the N-terminal region and the C-terminal region.
Furthermore, when considering the full-length S coding region, a marked aa substitution difference was observed between group I and group II (1.45 vs. 0.55, for substitutions per 100 aa, the same below, p=0.000) (Table 3). Moreover, the percentage of aa substitutions in the MHR for group I was significantly higher compared to those of group II (2.67 vs. 0.67, p=0.000) (Table 3). Those substitutions were strikingly found in the “a” determinant (4.12 vs. 1.47, p=0.022) (Table 3). Interestingly, great differences were also found in the N-terminal region between the two groups (1.01 vs. 0.42, p=0.044). These results indicate that the coexistence of HBsAg and anti-HBs is related to the accumulation of mutated residues, mostly, but not only, within the “a” determinant.
Mutation sI126S/T within the “a” determinant was the most frequent change, being present in 40% of HBV sequences from group I compared to 18% of HBV sequences from group II. In addition, only patients from group I had aa changes sQ129R, sG130N, sF134I, and sG145R, which have been proven to alter immunogenicity.4,16 However, substitution sG145R, which was popular in previous study, was only found in one patient from group I.
HBV reverse transcriptase sequencesAs the RT regions overlap the S gene, mutations within S gene might lead to aa substitutions in RT regions. Theoretically, the highest aa variability regions within the RT should be those overlapping the MHR, and especially the “a” determinant located in a structural and functional “finger” subdomain of the enzyme (Table 3).17 It was natural that the RT overlapping region the MHR, the number of aa substitutions of group I patients was markedly increased compared to those of group II patients (1.14 vs. 0.17, p=0.003). Surprisingly, in the RT overlapping region the “a” determinant was not significantly different in terms of frequencies of aa substitutions between groups I and II (2.22 vs. 0.49, p=0.073). When considering the region of RT overlapping the C-terminal region, which stands within the structural and functional “palm” subdomain of the enzyme,17 the difference of aa substitutions was not significant between the two groups (0.82 vs. 0.52, p=0.425). Furthermore, substitutions rtE134V, rtR138K, rtN139Q, corresponding to sI126S, sG130N, sT131N, were more frequent in patients from group I than in patients from group II.
DiscussionThe present study described the prevalence of coexistence of anti-HBs in HBsAg-positive CHB patients infected with genotype C HBV and analyzed the clinical and virological features, including aa substitutions in S gene and the overlapping RT regions. The presence of both HBsAg and anti-HBs was documented in 3.0% of 1194 CHB patients, which was similar to several previous reports,12,17–20 but lower compared to others.4,21 The prevalence diversity might be explained by differences in demographic, ethnic, geographical diversion, and diagnosis criteria. In this study, only an anti-HBs titer>10mIU/mL was considered positive according to the Chinese consensus criteria.22
Genotypes B and C are the most common HBV genotypes in China, accounting for approximately 95% of HBV-infected patients.23 Previous reports indicated that HBV genotype C takes more aggressive clinical course than genotype B.7,24 In addition, some studies suggested that concomitant HBsAg/anti-HBs is associated with more advanced liver diseases, such as fibrosis, cirrhosis, and hepatocellular carcinoma.21,25,26 Therefore, it is necessary to explore the mechanism underlying the coexistence of HBsAg and anti-HBs in genotype C CHB patients.
The mechanism underlying the simultaneous HBsAg/anti-HBs positivity is still unknown, but one explanation might be the selection of HBsAg immune escape variants. Several studies reported that, for the MHR, especially for the “a” determinant region, genetic variability in patients carrying concurrent HBsAg/anti-HBs is much higher than that in patients carrying HBsAg only.4,16,18,19,27 It has been reported that the accumulation of aa substitutions in and around the “a” determinant region may change the conformational structure and immunogenicity of HBsAg and T-cell epitope structure, leading to escape from recognition by the host immune system.13,14 This hypothesis is strengthened by the results of our study.
In the present study, a significantly higher aa variability was also observed within the MHR and the “a” determinant region of HBV from HBsAg+/anti-HBs+ patients than from controls, which is in line with previous reports.4,16,18,19,27 Whether and how these mutations affect the immunogenicity of HBsAg, either alone or in combination, still needs much more in-depth studies. However, regarding the specific aa substitutions, mutations sI126S/T, sQ129R, sG130N, sF134I, and sG145R found in this study were associated with alteration of HBsAg antigenicity, as reported previously.4,16,18,19,27 Interestingly, the most frequent substitution sG145R reported in previous studies was only found in one patient from group I.4,19 This result is similar to several previous reports from China.16,18,27 Diversities in genotypes may be an important reason for the preferred mutations in the “a” determinant. All patients enrolled in this study and most of those enrolled in other studies from China were infected with genotype C HBV, in contrast with genotypes A and D HBV prevalent in Europe and the US.
In this study, we also observed significantly higher aa variability within the RT region of HBV from group I than from group II. Furthermore, we specifically observed significantly higher aa variability in the RT region overlapping the MHR region of HBsAg. However, in the RT region overlapping the C terminal region of HBsAg, which is associated with the emergence of viral resistance, there was no significant difference in the percentage of aa substitutions between group I and group II. This result is in line with one report,16 but contradicts another study.19 Although the virological and clinical implications of our findings remain unknown, some mutations in RT region have already been confirmed to be associated with specific clinical outcomes previously. Mutation rtM204V, observed in one patient in our study, was previously found to partially lead to the production and intracellular retention of truncated HBsAg forms.28 In addition, previous study has demonstrated that mutations rtL180M and rtM204V confer Lamivudine resistance.29 Furthermore, we observed that the substitution rtS78T, which was found in group I, was previously found to decrease RT binding affinity of adefovir and potentially contributes by itself to adefovir resistance.30 Besides, mutation rtS78T, corresponding to a stop-codon at the HBsAg-position 69, might potentially affect HBV pathogenicity and oncogenic potential.31 Therefore, further studies deserve careful follow-up of these patients once antiviral treatments are introduced.
Notably, though most of previous reports supported the relationship of the HBsAg sequence variations and the concurrent HBsAg/anti-HBs serological profile, another study objected. In that study, similar aa variability was found in the HBsAg sequences derived from patients with and without anti-HBs.12 They thought the exposure of patients to HBV infection of different subtypes might be a possible explanation.12
In summary, our study has shown the prevalence of anti-HBs in HBsAg-positive CHB patients infected with genotype C HBV. We observed a higher aa variability within the N-terminal region and the MHR, especially the “a” determinant region, and within RT for regions overlapping the MHR in patients with concurrent HBsAg/anti-HBs serological profile. Additional research on these genetic mutants is needed to clarify their biological significance for viral persistence. Further studies are needed to determine whether these simultaneous HBV mutants might positively influence the clinical course and alter nucleos(t)ide analogs efficacy.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by the National Natural Science Foundation of China (81272693).