Disposal of Umbilical Cord Blood Units due to microbial contamination is a major problem in Cord Blood Banks worldwide as it reduces the number of units available for transplantation. Additionally, economic losses are generated as result of resources and infrastructure used to obtain such units. Umbilical Cord Blood Units that showed initial microbial contamination were subject to strains isolation, identification, and characterization by sequencing the 16S rRNA gene and Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR). Moreover, tests of antimicrobial resistance/sensitivity and phenotypic activities that may play an important role in microbial infection were performed. Microbial contamination was detected in 120 Umbilical Cord Blood Units (2.31%) in the period from 2003 to 2013. The most frequently isolated strains were Enterococcus faecium, followed by Staphylococcus epidermidis, Escherichia coli, Enterococcus faecalis, Staphylococcus haemoliticus, Klebsiella pneumoniae, Enterococcus durans, Lactobacillus helveticus, Enterococcus hiriae and Roseomonas genomospecies 5. The ERIC-PCR assays revealed a wide genetic diversity in some strains although belonging to the same genus and specie, indicating different sources of contamination. Broad-spectrum penicillins, third generation cephalosporins, aminoglycosides, and fluoroquinolones showed lower inhibitory activity on the tested strains. All strains were proteolytic, 67.69% were amylase-positive, 27.6% hemolysis-positive, and 34.71% nuclease-positive. The most common sources of contamination were: vaginal flora, digestive tract, and skin flora, highlighting the need for staff training in good manufacturing practices in collection SCU since all contaminants identified are part of the microbial flora of the donors. Implications and consequences in the therapeutic use of Umbilical Cord Blood Units for transplantation contaminated by multiresistant bacteria in immunocompromised patients are discussed.
In order to ensure Umbilical Cord Blood Units (UCBU) supply and safety, suitable for transplantation, it is necessary to have strict quality controls. These controls include: CD34+ cell count, typing by Human Leukocyte Antigen (HLA), serological tests, clonogenic capacity, and microbiological monitoring.1–3 Collection, manipulation, cryopreservation, and transplantation of UCBU involve a large number of procedures that are carried out in different areas and can result in microbial contamination of the final UCBU. For the most part, the collection of Cord Blood (CB) is carried out in an operating room with the use of safety procedures such as skin disinfection, closed collection systems, and sterile equipment among others. However, microbial contamination can occur during this process. Besides manipulation, cryopreservation, and thawing provide chances for the introduction of bacteria into UCBU.4 Additionally, several sources of contamination have been described, e.g. vaginal flora, skin, poor aseptic techniques in the areas, use of contaminated material, and others.5,6 It has been reported that the rate of contamination of UCBU ranges from 0 to 48%.7,8 The use of UCBU in sterile conditions is of vital importance because patients requiring this type of transplants are severely immunocompromised. Therefore, these patients have a high risk of contracting infections associated with the transplant. Previous studies have shown that the use of contaminated UCBU is one of the causes of morbidity and mortality in these patients.9 The identification of sources of contamination is crucial, from the sampling of CB until its processing in the laboratory. In previous research, identification of the contaminants was performed by phenotypic characteristics, such as biochemical tests, serotypes, and antimicrobial susceptibility.10,11 More recently, molecular typing methods, such as Enterobacterial Repetitive Intergenic Consensus (ERIC), based on Polymerase Chain Reaction (PCR) “ERIC-PCR”,12,13,14 have replaced previous techniques allowing better epidemiological determination of contamination sources. This molecular method is fast, simple, and highly reproducible. It also has been widely used in a large number of microbial types as well as in the study of clonality and identification of contamination sources. In addition, the new molecular technique for detecting bacteria allows for a quick and easy identification, compared with the classical microbiological methods. In this study we implemented the use of 16s rRNA gene sequencing and ERIC-PCR for typing the microbial strains obtained from criopreservated UCBU in the CB bank (CBB) of the National Center of Blood Transfusion (NCBT) in a period of 11 years (2003–2013) to identify potential sources of microbial contamination and carry out measures for their prevention. Tests of antimicrobial resistance/susceptibility and phenotypic activities that may play an important role in microbial infection were performed for the isolated strains. Implications and consequences of the therapeutic use of UCBU for transplantation contaminated by multiresistant bacteria in immunocompromised patients are discussed.
Materials and methodsA retrospective analysis over 11 years of cryopreserved UCBU in the CBB of the CNTS was performed. A total of 120 UCBU known to be contaminated by bacteria (aerobic, anaerobic, or both) were identified and extracted from the tank of liquid nitrogen (Bioarchive™ System TG 3626). They were immediately immersed in water bath at 37°C for thawing.
Enrichment of microbial contaminants in the BacT/ALERT 3D automated systemAll sampling was carried out under aseptic conditions in a laminar flow cabinet. All content (25mL) of each UCBU thawed was obtained by puncture with a hypodermic syringe. 12.5mL of the UCBU content were directly inoculated into the BacT/ALERT 3D: FA (FAN aerobic) and FN (anaerobic FAN) bottles (bioMérieux, Nueringen, Germany). Negative controls were established by inoculating 2mL of UCBU previously tested negative into BacT/ALERT 3D bottles. Over the seven-day incubation period, samples that exhibited a positive signal of contamination by the unit monitor of the BacT/ALERT 3D system were included in the study.
Isolation of microbial strainsBottles that showed positive signals of contamination were subcultered in solid media: 5% sheep blood agar, chocolate agar, Eosin Methylene Blue agar (EMB), mannitol salt agar, Pseudomonas agar, Trypticase Soy Agar (TSA), and Sabouraud Dextrose Agar (SDA). The plates were incubated aerobically at 37°C for 24–48h and at 28°C for 24–72h (only for SDA plates). The gas-pack system (BD GasPakTM EZ Gas Generating System) was used in the production of an anaerobic environment for the isolation of anaerobic bacteria in blood agar and in chocolate agar at 37°C for 48–72h. Subsequently, microbial strains were purified in the LB agar. All strains were cultured in LB-broth, then frozen in glycerol (50%) and, stored at −70°C. For molecular biology assays, total DNA from all strains was extracted as described using the QIAamp DNA Mini QIAcube Kit (QIAGEN, Germany).
Genetic identificationAll the amplification reactions were performed in a Touchgene Gradient thermal cycler Gene Amp® PCR System 9700 (Applied Biosystems). Polymerase chain reactions of the 16s rRNA gene were performed with universal primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) using the conditions recommended by DeSantis et al.15 Amplicons were analyzed on horizontals 1% agarose gels using 1× Tris–Borate–EDTA buffer (TBE), purified and sequenced by the Biology Institute, Universidad Nacional Autónoma de México (UNAM) using an ABI PRISM® 310 Genetic Analyzer sequencer (Applied Biosystems, California USA). Nucleotide sequences were compared with the nucleotide sequence database (GenBank) by means of the Blast algorithm (http://blast.ncbi.nlm.nih.gov).
Molecular typing by ERIC-PCRERIC-PCR assays were employed for typing all strains using the primers ERIC1R (5′-ATG TAA GCT CCT GGG GAT TCA-3′) and ERIC2 (5′-AAG TAA GTG ACT GGG GTG AGC G-3′). The total reaction volume was 50μL and consisted of Molecular Biology grade water, 1× PCR buffer, 20nM MgCl2, 25mM deoxyribonucleotide phosphate, 100pM of each primer, 3 units Taq DNA polymerase (Thermo Scientific) and 300ng of template DNA. Cycling conditions were as follow: pre-denaturation at 95°C for 7min, denaturation at 90°C for 30s, annealing at 58°C for 1min, and extension at 65°C for 8min, with a final extension at 68°C for 16min at the end for 30 cycles. Genetic profiles were run 1× TBE buffer, pH 8.3 and separated on horizontal electrophoresis in 1% agarose gels, visualized, photographed under UV illumination and analyzed by intra-gel pattern comparison.16
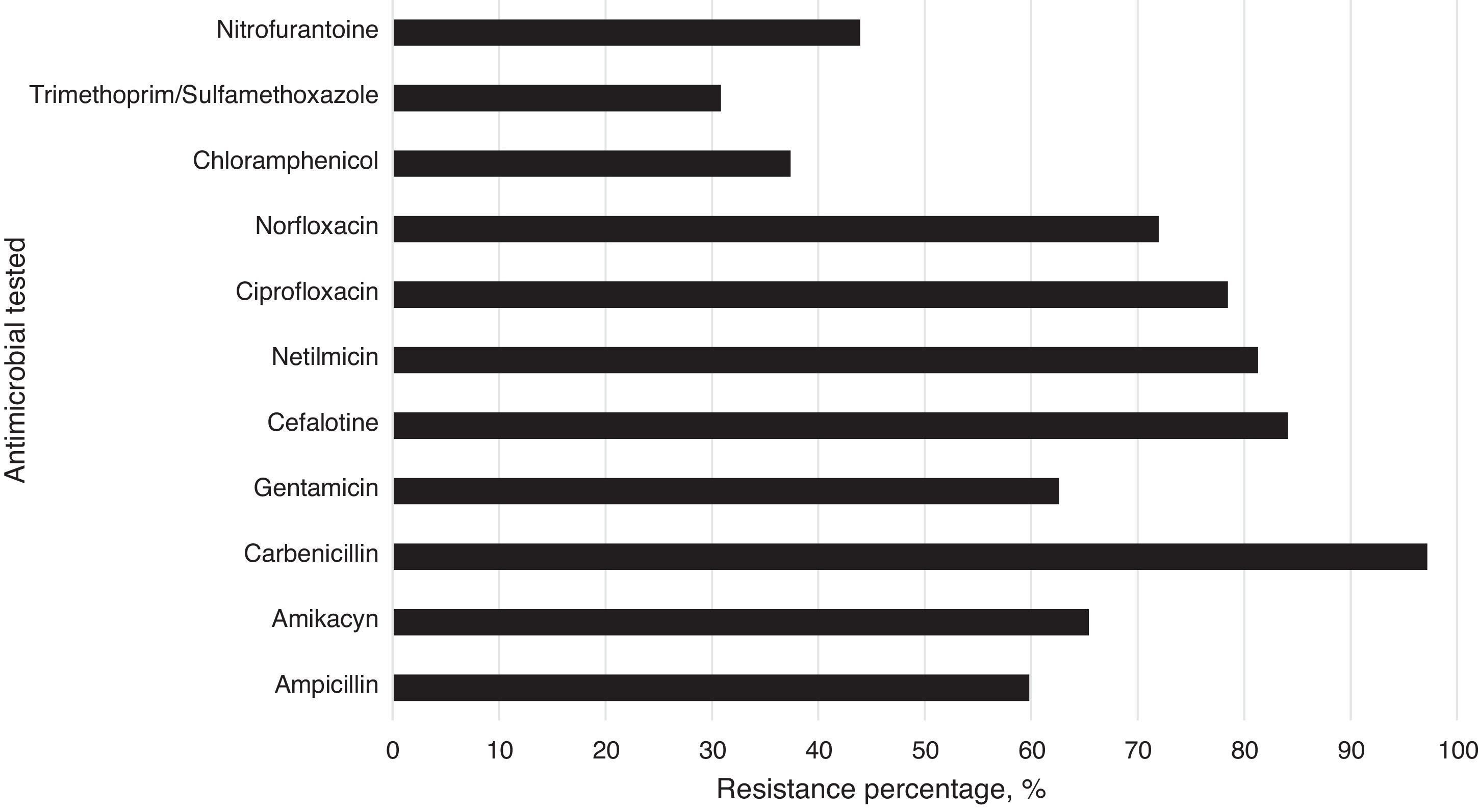
Susceptibility/resistance assaysResistance/susceptibility of all strains to different antibiotics were determined using the disk diffusion method on Mueller–Hilton agar plates according to recommendations of the clinical and laboratory standards institute (CLSI).17 The antibiotics used were: amikacyn (30μg), ampicillin (30μg), carbenicillin (100μg), cefalotine (30μg), cefotaxine (30μg), ciprofloxacin (5μg), chloramphenicol (30μg), gentamicin (10μg), netilmicin (30μg), nitrofurantoine (300μg), and trimethoprim/sulfamethoxazole (25μg). Antibiotics used in these tests are those that are commonly prescribed prophylactic drugs against Gram positive and negative bacteria. Pseudomonas aeruginosa ATCC 25923, Escherichia coli ATCC 25922, and S. aureus ATCC 25923 were used as controls. Results were interpreted as susceptible or resistant by measuring the diameter of inhibition zone according to the criteria stipulated by the CLSI. The frequency of antibiotic resistance was calculated and represented in percentages (%).
Phenotypic determination of virulence factorsThe strains were grown on TSA and two colonies were suspended in 2mL of LB-broth. The density of cellular suspensions was adjusted to 1×109CFU/mL in a spectrophotometer 3000 [SmartSpec™ Plus (BIO-RAD)] at 600nm. Cell suspensions were placed on various substrates by dripping for the phenotypic determination of virulence factors. All strains were tested in triplicate.
Proteolytic activityCasein hydrolysis was tested on Mueller–Hinton agar containing 10% (w/v) skimmed milk at 37°C for 24h. The presence of a transparent zone around the colonies indicated protease activity.
Amilolytic activityStarch hydrolysis was tested on LB agar (0.5%) supplemented with soluble starch (2%) at 37°C for 24h. The hydrolysis was demonstrated by exposure of the plates to iodine vapors for 5min. The presence of a transparent zone around the colonies indicated amylase activity.
Nuclease activityExtracellular nucleases (DNases) were determined on DNase agar plates (Difco) with 0.005% methyl green. The plates were incubated at 37°C for 24h. A pink halo around the colonies indicated nuclease activity.
Hemolytic activityThe strains were tested for hemolytic activity on agar base supplemented with 5% sheep erythrocytes. Fifty microliters of each cellular suspension were placed onto the plates and incubated at 37°C for 24h. The presence of a clear zone surrounding the colonies indicated hemolytic activity.
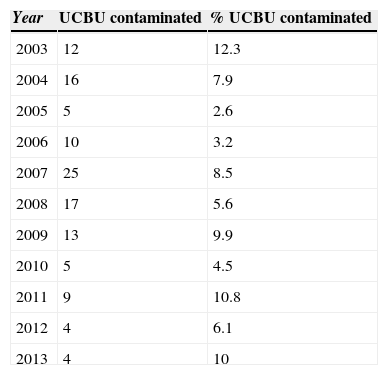
ResultsEnrichment of bacterial contaminants in the BacT/ALERT 3D automated systemA total of 5193 UCBU were collected in the NCBT cord blood bank. One hundred twenty were positive for microbial contamination (2.31%). Table 1 shows the contamination rate of UCBU in each year from 2003 to 2013. Microbial contamination was detected after thawing in 107 (89.16%) of 120 contaminated cryopreserved UCBU; in 13 (10.84%) UCBU bacterial contamination could not be confirmed after thawing. In 92 (85.98%) units just the aerobic bottles tested positive whereas 6 (5.6%) units grew in only the anaerobic bottles. The remaining 9 (8.4%) units both aerobic and anaerobic bottles tested positive. The mean time of detection of an initial positive culture was 17.9h for aerobic bottles and 36.2h for post-incubation anaerobic bottles. Regular microbiology techniques were performed to obtain pure cultures for genetic identification of isolates.
Annual contamination numerical and percentage rate of the Umbilical Cord Blood Units (UCBU) from 2003 to 2013. Distribution of the 120 contaminated UCBU of a total of 5193 UCBU in this period.
| Year | UCBU contaminated | % UCBU contaminated |
|---|---|---|
| 2003 | 12 | 12.3 |
| 2004 | 16 | 7.9 |
| 2005 | 5 | 2.6 |
| 2006 | 10 | 3.2 |
| 2007 | 25 | 8.5 |
| 2008 | 17 | 5.6 |
| 2009 | 13 | 9.9 |
| 2010 | 5 | 4.5 |
| 2011 | 9 | 10.8 |
| 2012 | 4 | 6.1 |
| 2013 | 4 | 10 |
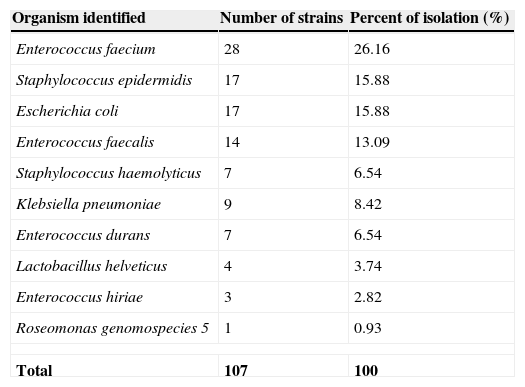
Genetic identification consisted in PCR amplification assays of V1–V9 regions (1450bp) of the 16s rRNA gene. The PCR products were purified, sequenced and compared to those of the GenBank database using strict filter parameters with more than 99% nucleotide homology and at least 80% query coverage. The sequences revealed a broad diversity of microorganisms phylogenetically different. As shown in Table 2, the most frequent families of isolated organisms belong to Enterococcaceae, Staphylococcaceae, Enterobacteriaceae, Lactobacillaceae and Acetobacteriaceae. Strains genetic identification of genus and species resulted in: Enterococcus faecium (28; 26.16%), followed by Staphylococcus epidermidis (17; 15.88%), Escherichia coli (17; 15.88%), Enterococcus faecalis (14; 13.08%), Klebsiella pneumoniae (9; 8.41%), Staphylococcus haemoliticus (7; 6.54%), Enterococcus durans (7; 6.54%), Lactobacillus helveticus (4; 3.73%), Enterococcus hiriae (3; 2.81%) and Roseomonas genosmoespecies 5 (1; 0.93%).
Isolation strains bacterial percentage obtained from 107 contaminated Umbilical Cord blood (UCB) units. Only the isolates obtained post-thaw are shown (107 UCBU).
| Organism identified | Number of strains | Percent of isolation (%) |
|---|---|---|
| Enterococcus faecium | 28 | 26.16 |
| Staphylococcus epidermidis | 17 | 15.88 |
| Escherichia coli | 17 | 15.88 |
| Enterococcus faecalis | 14 | 13.09 |
| Staphylococcus haemolyticus | 7 | 6.54 |
| Klebsiella pneumoniae | 9 | 8.42 |
| Enterococcus durans | 7 | 6.54 |
| Lactobacillus helveticus | 4 | 3.74 |
| Enterococcus hiriae | 3 | 2.82 |
| Roseomonas genomospecies 5 | 1 | 0.93 |
| Total | 107 | 100 |
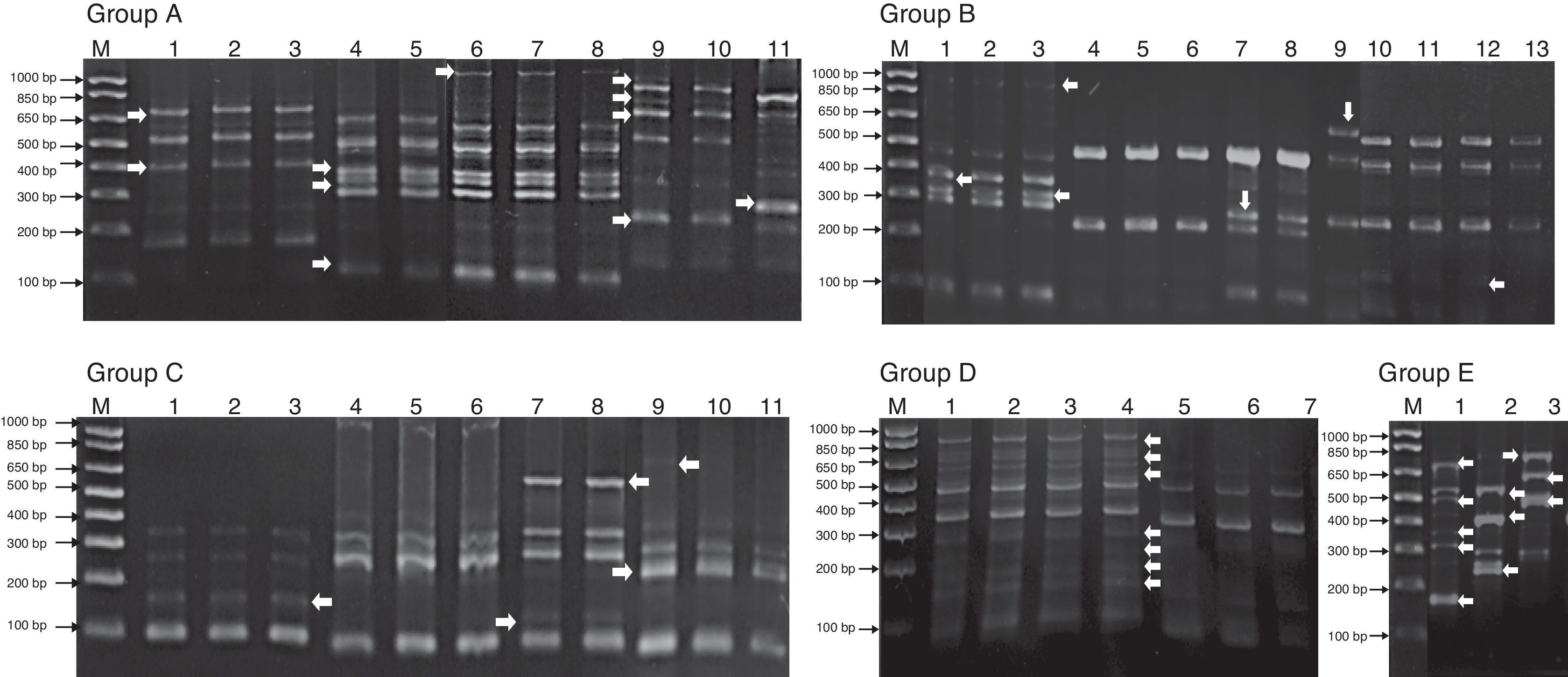
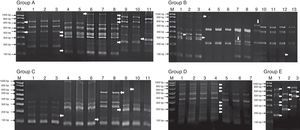
Genomic diversity analysis of 107 strains was carried out using ERIC-PCR fingerprinting method with ERIC-type primers (ERIC1R and ERIC2). The electrophoretic profiles of the DNA products obtained after PCR amplification using specific primers for ERIC sequences were determined for all strains obtained from contaminated UCBU. The electrophoretic analysis of PCR reaction products (amplicons) of the evaluated strains revealed that the number of bands ranged from 2 to 12 in different profiles. The sizes of the amplicons ranged from slightly more than 100bp to about 1100bp. Products in the range of 300–850bp were found more frequently. The genetic identification and ERIC-PCR profiles allowed differentiation of 107 strains which were clustered in 10 groups (Fig. 1) (Group A: E. durans strains; Group B: E. faecium strains; Group C: S. haemolyticus strains; Group D: S. epidermidis strains and Group E: K. pneumoniae strains). ERIC-PCR profiles of groups F, G, H, I, and J corresponds to E. hiriae, L. helveticus, Roseomonas genomospecies 5, E. fecalis, and E. coli respectively (not shown). Arrows (Fig. 1) indicate the intergenic variations in the genomes of strains of the same genus and species indicating a different contamination sources.
Bacterial representative ERIC-PCR fingerprints of isolated strains from Umbilical Cord Blood Units (UCBU). Group A: Enterococcus durans strains; Group B: Enterococcus faecium strains; Group C: Staphylococcus haemolyticus strains; Group D: Staphylococcus epidermidis strains and Group E: Klebsiella pneumoniae strains. M: 1000bp DNA marker (INVITROGEN). Arrows indicate the intergenic variations in the strains genomes of the same genus and species, indicating different contamination sources of UCBU.
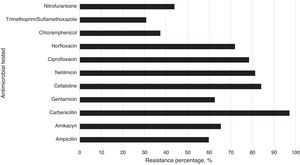
The isolated strains showed differences in susceptibility and resistance to the antimicrobials tested. These results show that sulfonamides, folate antagonists, nitrofurans, and first generation cephalosporins were the drugs with the best antimicrobial activity against all strains. The broad-spectrum penicillins, third generation cephalosporins, aminoglycosides, and fluoroquinolones showed lower inhibitory activity on the tested strains (Fig. 2).
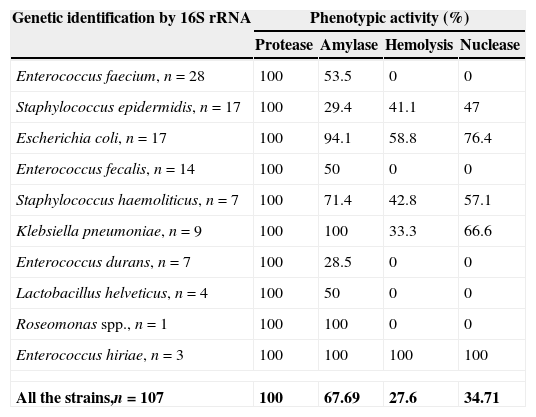
Phenotypic determination of virulence factorsResults of the phenotypic activity on different substrates are shown in Table 3. All strains showed protease activity while amylase, hemolytic-activity, and nuclease-activity varied among the strains tested.
Incidence of phenotypic expression (%) in bacterial strains isolated from Umbilical Cord Blood Units (UCBU).
| Genetic identification by 16S rRNA | Phenotypic activity (%) | |||
|---|---|---|---|---|
| Protease | Amylase | Hemolysis | Nuclease | |
| Enterococcus faecium, n=28 | 100 | 53.5 | 0 | 0 |
| Staphylococcus epidermidis, n=17 | 100 | 29.4 | 41.1 | 47 |
| Escherichia coli, n=17 | 100 | 94.1 | 58.8 | 76.4 |
| Enterococcus fecalis, n=14 | 100 | 50 | 0 | 0 |
| Staphylococcus haemoliticus, n=7 | 100 | 71.4 | 42.8 | 57.1 |
| Klebsiella pneumoniae, n=9 | 100 | 100 | 33.3 | 66.6 |
| Enterococcus durans, n=7 | 100 | 28.5 | 0 | 0 |
| Lactobacillus helveticus, n=4 | 100 | 50 | 0 | 0 |
| Roseomonas spp., n=1 | 100 | 100 | 0 | 0 |
| Enterococcus hiriae, n=3 | 100 | 100 | 100 | 100 |
| All the strains,n=107 | 100 | 67.69 | 27.6 | 34.71 |
This study reports the type and incidence of bacterial contamination of UCBU for transplant at the CBB of the National Center of Blood Transfusion in Mexico City in a period of 11 years (2003–2013). A total of 5193 CB samples were collected between May 2003 and December 2013. Of those, 1831 (35.25%) were accepted to be cryopreserved and 3, 362 (64.74%) units were excluded for various reasons: weight (suitable volume), white blood cells count (<7×106), total nucleated cell number (<8×108), CD34+ cell number (<2×106), expiration (>48h), doubly reactive serology (HCV, HBV, HIV, Chagas disease, brucellosis, or syphilis), and microbial contamination (aerobic, anaerobic, or both). The incidence of bacterial contamination of UCBU was 2.31%, or 120 units, of which in 13 units bacterial could not be recovered after thawing possibly due to bacterial thermal shocks following exposure to extreme temperatures. The bacterial contamination rate reported in this work are much lower than those reported in previous studies, with microbial contamination rates varying from 0 to 48%.7,8 In this work, a wide variety of bacterial contaminants were isolated and identified from the UCBU. The most frequent bacteria identified were associated with vaginal flora (3.73%), skin (22.44%), and gastrointestinal flora (70.87%). Roseomonas genomospecies 5 (0.93%) was also detected. The advantages of automated culture are: a shortened incubation period (down to 7 days) and rapid availability of positive results due to kinetic monitoring with continuous readout of CO2 production (BacT/ALERT System). However, this automated system focuses exclusively on the cultivation of mesophilic microorganisms, excluding potentially pathogenic bacteria and fungi such as Pseudomonas and Candida since their optimal growth temperature is 28°C. This is important when it comes to nosocomial bacteria that could contaminate CB and thus not be detected by these automated culture systems. Organisms detected in the UCBU were similar to those of other published reports.6,11,18 Gram-negative rods, members of the Enterobacteriaceae family are well known for their ability to cause life-threatening infections in humans. Although they do not normally colonize the skin, they may be present transiently. K. pneumoniae, Escherichia coli (both identified in this work), and other Enterobacteriaceae members have virulence factors such as toxins, adhesins, and capsules that may enable them cope with host defense mechanisms.19–22 Moreover, these organisms may have mobile genetic elements such as plasmids, integrons, and transposons that confer antibiotic resistance. Consequently, eradication of infections with common antimicrobial treatment in immunosuppressed patients become a relevant problem.23,24 This is the first study reporting isolation of Roseomonas genomospecies 5 from UCBU. This microorganism belongs to the Acetobacteriaceae family, is a Gram-negative coccobacilli that develops pink-pigmented colonies. Roseomonas are commonly isolated from middle-age women with one of several underlying conditions, including cancer and diabetes. They are also isolated from blood in association with clinical signs of sepsis (wounds).25 Although this genus of bacteria is not considered a primary human pathogen, it has been isolated as commensal bacteria from immunosuppressed patients and young adults with sexually transmitted diseases.26 Presumably, this organism is associated with cross-contamination in the operating room or infection in the CB donor who could be immunosuppressed and unware of it. Additionally, Gram-positive bacteria such as Staphylococcus spp. and Enterococcus spp. are known to survive after cryopreservation processes due to cell wall characteristics. E. faecalis, is a perianal contaminant commonly associated with the environment during CB collection.27 Poor aseptic techniques could be the reason for the high incidence of this bacterial contaminant in this study. Contamination of UCBU is influenced by multiple factors such as bad collection techniques, exhaustive manipulation of CB, CBB size, and others. In this work, the samples of CB were most likely contaminated during collection, due to inadequate skin aseptic technique, direct contact with fecal bacteria during childbirth, and contact with the vaginal flora. There are several sources of CB contamination because they are collected using different methods. Additionally, childbirth (cesarean or normal) is directly related to the sterility of the UCBU. Skin, equipment of collection, processing, and laboratory equipment such as water baths, incubators, centrifuges, and nitrogen tanks have been shown to be potential sources of contamination.6,8,28 Microbial contamination of CB has been significantly reduced in the past eight years in the CBB, as a result of strategies including disinfection methods, automated detection of microbial contaminants, and others (Table 1). These strategies have been successful because they aimed at reducing microbial contamination originated from regular vaginal biota, enteric and transient skin flora. In order to identify clonal relationship between isolates of the same genus and species, molecular typing assays (ERIC-PCR) were performed. Contamination sources were clearly identified in this work in different sites because even if the identified bacteria belonging to the same genus and species, they presented intergenic variations according to molecular typing assays (Fig. 1). Arrows in the different profiles indicate the intergenic variations in the strains genomes of the same genus and species indicating different contamination sources of UCBU. The use of molecular typing techniques such as RAPD-PCR, ERIC-PCR, DGGE-PCR, and others might identify the source of contamination during CB collection and processing distinguishing transient contaminations from intrinsic contamination and undetected bacteremia in the donor. This molecular tool has been widely used to rule out clonal relationships between bacteria from the same genus and species isolated from a specific area.29,30 Correlation of the antimicrobial resistance pattern and plasmid profiles with clonality in contaminants strains is limited, especially in the hospital environment. Here the use of antibiotics generates great selective pressure for resistance acquisition by means of mobile genetic elements such as plasmids, transposons, integrons, and others. A high proportion of strains resistant to first, second, and third generation antibiotics were identified in this work. Previous work has reported the use of UCBU contaminated with bacteria and recommended administration of antibiotics prior to transplantation. Nevertheless, the risk of infection associated with multi-resistant bacteria to antibiotics persists. Patterns of antibiotic resistance found in isolated strains clearly show molecular mechanisms directly involved in the resistance phenotype. It is well known that prior to UCBU transplantation; the patient who receives antimicrobial treatment and immunosuppressive therapy remains with a potential risk of infection, as the antibiotic therapy may not be effective against multidrug resistant contaminants. Further investigations should aim to elucidate the molecular mechanisms involved in antibiotic resistance in isolates of UCBU isolated in this work. The production of exoenzymes and hemolysins are defense mechanisms of microorganisms during their colonization and pathogenesis. The presence of hemolysins, proteases, amylases, and DNases in strains isolated in UCBU clearly shows the potential risk associated to phenotypically identified virulence factors. Molecular assays have also been used to identify virulence factors.31 Although the incidence of contaminated UCBU found in this work is low, it is recommended to systematically carry out quality control protocols in order to prevent microbial contamination (obtaining the CB in the operating room, CBB processes, and cryopreservation). Recently, a measure to prevent microbial contamination of skin CB associated bacteria has been implemented. This technique involves the deviation and disposal of the first milliliters of CB to eliminate the possibility of entering bacteria into the collection bag. Although there are standardized protocols for the collection, transport, processing, and cryopreservation of CB, it is extremely important to continue with Good Manufacturing Practices that include all of the procedures to obtain UCBU. The need to have a wide genetic diversity of UCBU for transplantation in our country is of great importance and the presence of contaminated UCBU directly affects the genetic diversity of UCBU of CBB.
Conflicts of interestThe authors declare no conflicts of interest.