In this study, 922 consecutive non-duplicate clinical isolates of Enterobacteriaceae obtained from hospitalized and non-hospitalized patients at Bejaia, Algeria were analyzed for AmpC-type β-lactamases production. The ampC genes and their genetic environment were characterized using polymerase chain reaction (PCR) and sequencing. Plasmid incompatibility groups were determined by using PCR-based replicon typing. Phylogenetic grouping and multilocus sequence typing were determined for molecular typing of the plasmid-mediated AmpC (pAmpC) isolates.
Of the isolates, 15 (1.6%) were identified as AmpC producers including 14 CMY-4-producing isolates and one DHA-1-producing Klebsiella pneumoniae. All AmpC-producing isolates co-expressed the broad-spectrum TEM-1 β-lactamase and three of them co-produced CTX-M and/or SHV-12 ESBL. Phylogenetic grouping and virulence genotyping of the E. coli isolates revealed that most of them belonged to groups D and B1. Multilocus sequence typing analysis of K. pneumoniae isolates identified four different sequence types (STs) with two new sequences: ST1617 and ST1618. Plasmid replicon typing indicates that blaCMY-4 gene was located on broad host range A/C plasmid, while LVPK replicon was associated with blaDHA-1. All isolates carrying blaCMY-4 displayed the transposon-like structures ISEcp1/ΔISEcp1-blaCMY-blc-sugE.
Our study showed that CMY-4 was the main pAmpC in the Enterobacteriaceae isolates in Algeria.
Infection with resistant organisms is a major public health issue. Enterobacteriaceae are important causes of both community-acquired and healthcare-associated infections in adults and children, and production of β-lactamases is of greater concern.1 Plasmid-mediated AmpC (pAmpC) β-lactamases have emerged and are being reported worldwide with varying prevalence rates.2 They have been mainly detected in Escherichia coli, Klebsiella spp., Salmonella spp., and Proteus mirabilis.3 These enzymes confer resistance to penicillins, first and third generation cephalosporins, cephamycins, and monobactams such as aztreonam. They are poorly inhibited by the commercially available β-lactamase inhibitors such as clavulanic acid, but are inhibited by cloxacillin and phenylboronic acid. Treatment options are severely limited because pAmpC are often associated with other multiple resistance genes, such as those of resistance to quinolones as well as other β-lactamase genes.3,4 The acquired ampC genes have emerged following mobilizations mediated by such elements as IS26, ISEcp1, or ISCR1.3 Thus, ISEcp1 has played an important role in mobilizing blaCMY-2-like genes, since it is often found at their 5′ flanks.5,6 It has been identified in plasmids of the Inc A/C and Inc I1 groups.5–7 ISCR1 elements have been found adjacent to a number of ampC genes, including blaDHA-1, as well as plasmid-mediated quinolone resistance determinant qnr.8 Generally, the blaDHA-1 gene has been mainly associated with Inc FII and Inc L/M plasmids.9
In Algeria, only few reports on plasmid-encoded AmpC (CMY-2 and DHA-1) in Enterobacteriaceae strains recovered from hospital settings were published.10–12
The aim of this study was to investigate the prevalence and molecular epidemiology of cefoxitin resistance among Enterobacteriaceae isolates recovered from hospitalized and non-hospitalized patients in Bejaia locality (Algeria). The association of pAmpC with extended-spectrum β-lactamase (ESBL) and plasmid-mediated quinolone resistance determinant was also studied.
Materials and methodsBacterial strainsA total of 922 non-duplicate isolates (one per patient) of Enterobacteriaceae were collected from March 2005 to April 2010 from the Microbiology Laboratories of five hospitals and four private laboratories in Bejaia (Algeria). The isolates were recovered from various pathological specimens and were identified by the API 20E system (bioMérieux, Marcy l’Etoile, France) as follows: E. coli (n=551); Klebsiella pneumoniae (n=221); P. mirabilis (n=125) and Salmonella sp. (n=25).
E. coli J53AzR was used as recipient strains for conjugation experiments. E. coli DH10B (Invitrogen) was used in transformation experiments and E. coli ATCC 25922 was used as a quality control strain for antimicrobial susceptibility testing.
Antimicrobial susceptibility testingAntibiotic susceptibility was determined on Mueller Hinton agar by standard disk diffusion procedure as described by the European Committee on Antimicrobial Susceptibility Testing (2014),13 for the following antibiotics: aztreonam, ticarcillin, piperacillin, amoxicillin–clavulanate, ticarcillin–clavulanate, cefoxitin, cefepime, piperacillin–tazobactam, cefuroxime, cefotaxime, ceftazidime, imipenem, tobramycin, amikacin, gentamicin, sulfonamide, trimethoprim, nalidixic acid, ciprofloxacin, norfloxacin, tetracycline, and chloramphenicol (BioRad, Marnes La Coquette, France). For tetracycline, the Antibiogram Committee of the French Society for Microbiology recommendations breakpoints were used (http://www.sfm-microbiologie.org).
Isolates showing a zone of inhibition diameter ≤20mm with cefoxitin were selected for screening for pampC genes. ESBL production was detected by a double-disk synergy test (DDST) on Mueller Hinton supplemented with cloxacillin (200mg/L).14 Inducibility of the β-lactamase was determined by the double disk test. The cephalosporins used were cefotaxime, ceftazidime, and cefepime. Clavulanic acid (10μg) and cefoxitin (30μg) were used as inducing agents. The plates were examined after overnight incubation at 37°C.15
Minimum inhibitory concentrations (MICs) of amoxicillin, amoxicillin/clavulanate, piperacillin/tazobactam, cefotaxime, ceftazidime, cefoxitin, imipenem, aztreonam, and cefepime were determined by Etest (AB bioMérieux, Marcy l’Etoile, France).
Molecular characterization of resistance determinantsTotal DNA was extracted by using a QIAmp DNA Mini Kit (QIAGEN) according to the instructions of the manufacturer. A multiplex PCR covering the six families of ampC genes (CMY-2/BIL/LAT, CMY-1/MOX, DHA, FOX, ACC, ACT/MIR) was performed as previously described.16 PCR-positive isolates were further tested using individual pairs of primers and then sequenced. pAmpC-producing isolates positive for the DDST were screened for blaCTX-M, blaTEM and blaSHV by PCR as described previously.17
Screening of qnrA, qnrB, qnrS, qnrC, qnrD and qepA genes was carried out with a multiplex real-time PCR assay using SYBR Green I and Roche LightCycler1 as described previously.18 Pyrosequencing method was used for the detection of aac(6′)-Ib-cr and aac(6′)-Ib genes.19
All PCR products were sequenced and the sequencing results were compared to reported sequences available in GenBank.
Transfer of resistanceConjugation was performed on Mueller Hinton agar supplemented with sodium azide (100mg/L) and cefotaxime (1mg/L). Transconjugants growing on the selection plates were subjected to antimicrobials susceptibility, DDST and PCR analysis to confirm the presence of the AmpC phenotype.
Molecular typingPossible genomic relatedness of strains was analyzed by RAPD using genomic DNA as previously described.20 Multilocus sequence typing (MLST) was performed on the K. pneumoniae isolates using seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB).21 A detailed protocol of the MLST procedure, including allelic type and ST assignment methods, is available in MLST databases from the Pasteur Institute, Paris, France, at the website http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html.
Phylogenetic groups and virulence genotyping of E. coliPCRs were performed to determine the phylogenetic groups (A, B1, B2, C, D, E, F, and clade I) of the E. coli isolates, using the newly revised Clermont method.22 All isolates belonging to group B2 were analyzed by two multiplex PCR as described previously.23 The presence of eight virulence factors found in ExPEC was investigated by PCR. These factors included sfa/foc (S and F1C fimbriae), papG alleles (G adhesin classes of P fimbriae), papC (C adhesin classes of P fimbriae), hlyA (alpha-haemolysin A), cnf (cytotoxic necrotizating factor 1), fyuA (genes of yersiniabactin), iutA (aerobactin receptor), and ibeA (invasion protein IbeA).
Plasmid replicon typingPlasmids incompatibility (Inc) groups were determined using PCR-based replicon typing (PBRT).24 Four multiplex PCR were used for the detection of A/C, T, FIIAs, W, N, FIB, L/M, I1-Iγ, X, HI2, FIA, and Y replicons. Replicons P, R, U, F, FIC, HI1, B/O and K were detected by simplex PCR.24,25 Replicons FII1K, FII2K, NewXXX also named ZK, LVPK, and Amet were detected using PCR method described by D. Decré and G. Arlet.
Genetic organization of blaampC genesFor the analysis of genetic arrangement of the resistance genes, overlapping PCR amplification of internal regions of the transposon-like element that carried blaCMY-4 was performed based on known sequences.
Genetic structures surrounding the blaDHA-1 gene were studied by PCR mapping, cloning and sequencing method using a large variety of primers based on the previously reported structures.8
The nucleotide sequence and the deduced protein sequence were analyzed using the Basic Local Alignment Search Tool (BLAST) through the Internet (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignment of deduced peptide sequences was carried out with the Vector NTI program (Invitrogen).
ResultsBacterial isolates and antibiotic susceptibilityAmong the 922 Enterobacteriaceae isolates, 15 isolates showed decreased susceptibility to cefoxitin: nine isolates of E. coli (9/551), five isolates (5/221) of K. pneumoniae and one isolate (1/125) of P. mirabilis. Ten isolates were recovered from urine. Six E. coli isolates and one K. pneumoniae isolate were from community patients (7/712), and the remaining eight isolates were collected from hospitalized patients (8/210).
All isolates exhibited resistance to ticarcillin, piperacillin, ticarcillin–clavulanic acid, amoxicillin–clavulanic acid, cefuroxime, cefotaxime, ceftazidime, aztreonam, and cefoxitin. Isolates exhibited intermediate resistance to piperacillin–tazobactam (60%) and cefepime (40%). Resistance of the isolates to non-β-lactam antibiotics was high for sulfonamide (80%), tobramycin (71.4%), gentamicin (71.4%), tetracycline (71.4%), chloramphenicol (64.3%) and trimethoprim (57.1%), and low for nalidixic acid (28.6%) and amikacin (6.6%). The isolates remain susceptible to imipenem and fluoroquinolones. The MICs ranges are listed in Table 1.
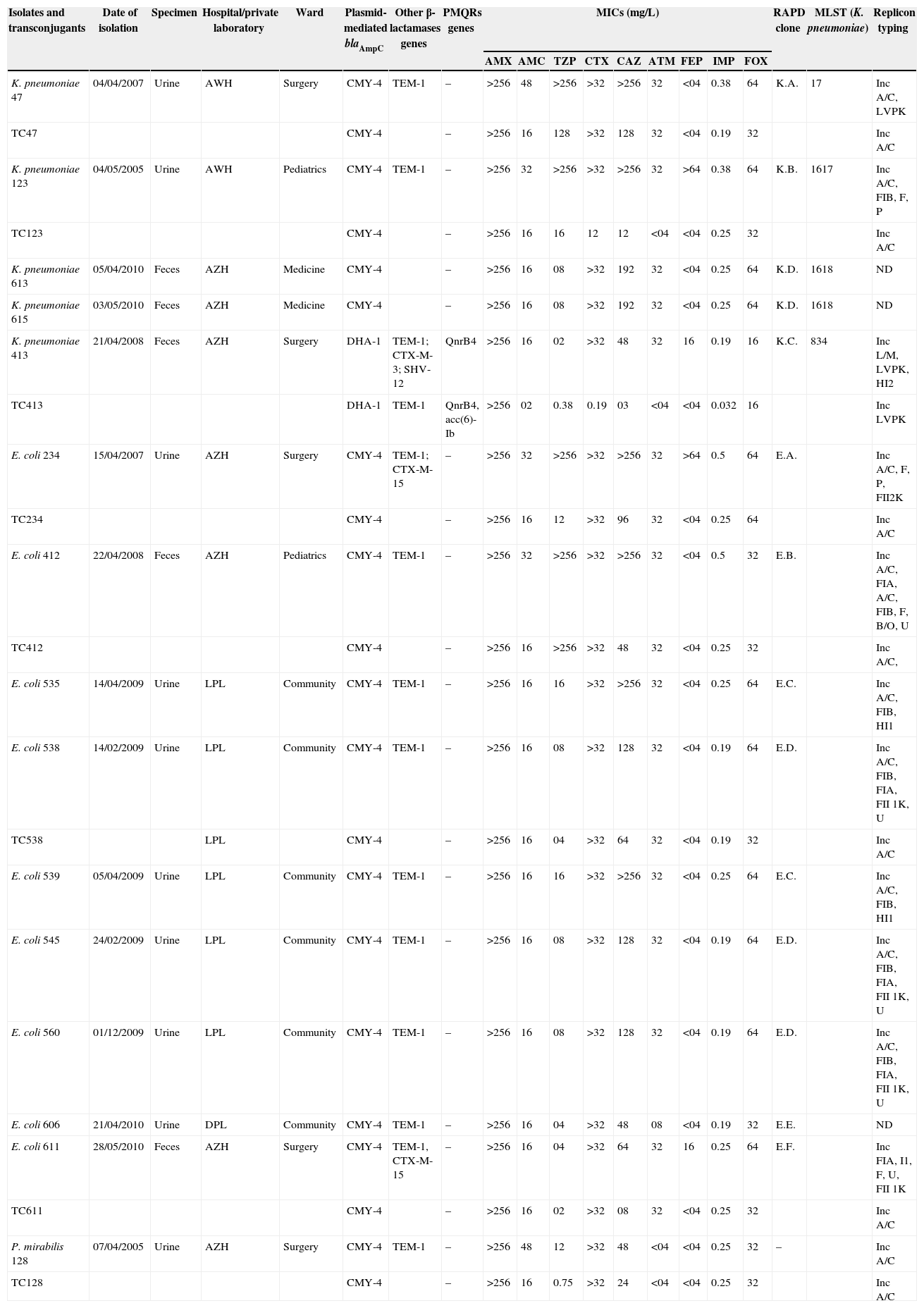
Microbiological features of pAmpC producing Enterobacteriaceae.
| Isolates and transconjugants | Date of isolation | Specimen | Hospital/private laboratory | Ward | Plasmid-mediated blaAmpC | Other β-lactamases genes | PMQRs genes | MICs (mg/L) | RAPD clone | MLST (K. pneumoniae) | Replicon typing | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | TZP | CTX | CAZ | ATM | FEP | IMP | FOX | |||||||||||
| K. pneumoniae 47 | 04/04/2007 | Urine | AWH | Surgery | CMY-4 | TEM-1 | – | >256 | 48 | >256 | >32 | >256 | 32 | <04 | 0.38 | 64 | K.A. | 17 | Inc A/C, LVPK |
| TC47 | CMY-4 | – | >256 | 16 | 128 | >32 | 128 | 32 | <04 | 0.19 | 32 | Inc A/C | |||||||
| K. pneumoniae 123 | 04/05/2005 | Urine | AWH | Pediatrics | CMY-4 | TEM-1 | – | >256 | 32 | >256 | >32 | >256 | 32 | >64 | 0.38 | 64 | K.B. | 1617 | Inc A/C, FIB, F, P |
| TC123 | CMY-4 | – | >256 | 16 | 16 | 12 | 12 | <04 | <04 | 0.25 | 32 | Inc A/C | |||||||
| K. pneumoniae 613 | 05/04/2010 | Feces | AZH | Medicine | CMY-4 | – | >256 | 16 | 08 | >32 | 192 | 32 | <04 | 0.25 | 64 | K.D. | 1618 | ND | |
| K. pneumoniae 615 | 03/05/2010 | Feces | AZH | Medicine | CMY-4 | – | >256 | 16 | 08 | >32 | 192 | 32 | <04 | 0.25 | 64 | K.D. | 1618 | ND | |
| K. pneumoniae 413 | 21/04/2008 | Feces | AZH | Surgery | DHA-1 | TEM-1; CTX-M-3; SHV-12 | QnrB4 | >256 | 16 | 02 | >32 | 48 | 32 | 16 | 0.19 | 16 | K.C. | 834 | Inc L/M, LVPK, HI2 |
| TC413 | DHA-1 | TEM-1 | QnrB4, acc(6)-Ib | >256 | 02 | 0.38 | 0.19 | 03 | <04 | <04 | 0.032 | 16 | Inc LVPK | ||||||
| E. coli 234 | 15/04/2007 | Urine | AZH | Surgery | CMY-4 | TEM-1; CTX-M-15 | – | >256 | 32 | >256 | >32 | >256 | 32 | >64 | 0.5 | 64 | E.A. | Inc A/C, F, P, FII2K | |
| TC234 | CMY-4 | – | >256 | 16 | 12 | >32 | 96 | 32 | <04 | 0.25 | 64 | Inc A/C | |||||||
| E. coli 412 | 22/04/2008 | Feces | AZH | Pediatrics | CMY-4 | TEM-1 | – | >256 | 32 | >256 | >32 | >256 | 32 | <04 | 0.5 | 32 | E.B. | Inc A/C, FIA, A/C, FIB, F, B/O, U | |
| TC412 | CMY-4 | – | >256 | 16 | >256 | >32 | 48 | 32 | <04 | 0.25 | 32 | Inc A/C, | |||||||
| E. coli 535 | 14/04/2009 | Urine | LPL | Community | CMY-4 | TEM-1 | – | >256 | 16 | 16 | >32 | >256 | 32 | <04 | 0.25 | 64 | E.C. | Inc A/C, FIB, HI1 | |
| E. coli 538 | 14/02/2009 | Urine | LPL | Community | CMY-4 | TEM-1 | – | >256 | 16 | 08 | >32 | 128 | 32 | <04 | 0.19 | 64 | E.D. | Inc A/C, FIB, FIA, FII 1K, U | |
| TC538 | LPL | CMY-4 | – | >256 | 16 | 04 | >32 | 64 | 32 | <04 | 0.19 | 32 | Inc A/C | ||||||
| E. coli 539 | 05/04/2009 | Urine | LPL | Community | CMY-4 | TEM-1 | – | >256 | 16 | 16 | >32 | >256 | 32 | <04 | 0.25 | 64 | E.C. | Inc A/C, FIB, HI1 | |
| E. coli 545 | 24/02/2009 | Urine | LPL | Community | CMY-4 | TEM-1 | – | >256 | 16 | 08 | >32 | 128 | 32 | <04 | 0.19 | 64 | E.D. | Inc A/C, FIB, FIA, FII 1K, U | |
| E. coli 560 | 01/12/2009 | Urine | LPL | Community | CMY-4 | TEM-1 | – | >256 | 16 | 08 | >32 | 128 | 32 | <04 | 0.19 | 64 | E.D. | Inc A/C, FIB, FIA, FII 1K, U | |
| E. coli 606 | 21/04/2010 | Urine | DPL | Community | CMY-4 | TEM-1 | – | >256 | 16 | 04 | >32 | 48 | 08 | <04 | 0.19 | 32 | E.E. | ND | |
| E. coli 611 | 28/05/2010 | Feces | AZH | Surgery | CMY-4 | TEM-1, CTX-M-15 | – | >256 | 16 | 04 | >32 | 64 | 32 | 16 | 0.25 | 64 | E.F. | Inc FIA, I1, F, U, FII 1K | |
| TC611 | CMY-4 | – | >256 | 16 | 02 | >32 | 08 | 32 | <04 | 0.25 | 32 | Inc A/C | |||||||
| P. mirabilis 128 | 07/04/2005 | Urine | AZH | Surgery | CMY-4 | TEM-1 | – | >256 | 48 | 12 | >32 | 48 | <04 | <04 | 0.25 | 32 | – | Inc A/C | |
| TC128 | CMY-4 | – | >256 | 16 | 0.75 | >32 | 24 | <04 | <04 | 0.25 | 32 | Inc A/C | |||||||
LPL, Lalaoui private laboratory; DPL, Djama private laboratory; AZH, Amizour hospital; AWH, Amriw hospital; TZP, piepracillin–tazobactam; AMX, amoxicillin; AMC, amoxicillin–clavulanic acid; FOX, cefoxitin; ATM, aztreonam; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; IMP, imipenem; ND, not determined.
The ESBL phenotypic screening by double disk diffusion synergy test showed that one isolate of K. pneumoniae and two isolates of E. coli were ESBL producers (Table 1).
Inducibility of β-lactamases was recognized by the disk antagonism test, which demonstrated blunting of the cephalosporin disks adjacent to the cefoxitin and clavulanic acid disks in only one isolate of K. pneumoniae 413. This phenotype suggested the presence of an inducible AmpC-type β-lactamase.
Genotypic analysis of antibiotic resistance genesBy multiplex PCR, we obtained amplicons in 15 isolates: nine E. coli isolates, five K. pneumoniae isolates, and one P. mirabilis isolate (Table 1).
PCR and sequencing analysis revealed the presence of blaCMY-4 in all isolates except one isolate of K. pneumonia, which produced blaDHA-1.
In addition, two E. coli (CMY-4) co-produced CTX-M-15 ESBLs and one isolate of K. pneumoniae (DHA-1) co-produced CTX-M-3 and SHV-12 ESBL. All isolates carried blaTEM-1.
PCR amplification of PMQR yielded amplification in one K. pneumoniae isolate only. This strain expressed both the qnrB4, aac(6′)-Ib, blaDHA-1, blaCTX-M-3, blaSHV-12 and blaTEM-1 genes (Table 1).
No amplicons were obtained for qnrA, qnrS, qnrD, qnrC and qepA in all isolates.
Conjugation and replicon typingBy mating assay, the ampC genes were transferred from three of the five K. pneumoniae, five of the nine E. coli isolates and from the P. mirabilis isolate. Susceptibility results of the transconjugants are shown in Table 1.
PBRT of the plasmid Inc groups showed that the plasmids carrying blaCMY-4 belonged to the Inc A/C group and the plasmid carrying blaDHA-1 belonged to the group Inc LVPK (Table 1).
Molecular typingRAPD-typing revealed the presence of diverse bacterial population and no predominant clone was identified in our collection.
MLST analysis of the five AmpC-producing K. pneumoniae identified four different STs, including ST17, ST834 and two new sequence types: ST1617 (Kp 123) and ST1618 (Kp 613 and 615) (Table 1). In ST1618, we described a new allele's mdh and rpoB designated respectively 145 and 108. The typing results generated by RAPD analysis among the isolates were compatible with those obtained by MLST.
E. coli phylogenetic groups and virulence factorsOf the nine E. coli isolates, three belonged to group D, three to group B1 (recovered from urine), two to group B2, and the last one to group F (Table 2).
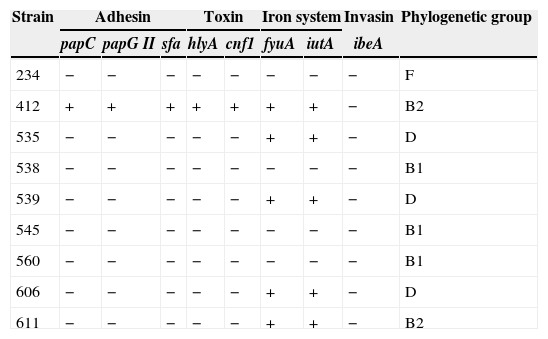
Distribution and combination patterns of virulence genes and phylogenetic groups detected in pAmpC-producing E. coli.
| Strain | Adhesin | Toxin | Iron system | Invasin | Phylogenetic group | ||||
|---|---|---|---|---|---|---|---|---|---|
| papC | papG II | sfa | hlyA | cnf1 | fyuA | iutA | ibeA | ||
| 234 | − | − | − | − | − | − | − | − | F |
| 412 | + | + | + | + | + | + | + | − | B2 |
| 535 | − | − | − | − | − | + | + | − | D |
| 538 | − | − | − | − | − | − | − | − | B1 |
| 539 | − | − | − | − | − | + | + | − | D |
| 545 | − | − | − | − | − | − | − | − | B1 |
| 560 | − | − | − | − | − | − | − | − | B1 |
| 606 | − | − | − | − | − | + | + | − | D |
| 611 | − | − | − | − | − | + | + | − | B2 |
Groups F and B1 were not found in any selected virulence genes.
Five isolates harbored genes encoding siderophores (fyuA, iutA).
The E. coli 412 isolate was assigned to the B2 sub-group VII and STc14. This strain was isolated from the feces of a hospitalized patient. This isolate contained a blaTEM-1 gene and, a blaCMY-4 gene, which was transferred with an Inc A/C plasmid. The following virulence genes (papC, papG II, sfa, hlyA, cnf1, fyuA and iutA) were detected in this isolate (Table 2).
By using allele-specific PCR method for detecting the main E. coli B2 STc, E. coli 611 isolate was unassigned; it did not give any PCR products except for the internal control.
Characterization of the genetic contexts of blaAmpC genesAnalysis of the genetic structure of the blaCMY-4 gene in our collection showed that it was located on a transposon-like DNA element consisting of a specific ISEcp1/ΔISEcp1-blaCMY-4-blc-sugE structure. This structure was similar to that found in plasmid pCC416 (GenBank AJ875405).
A region typical of a complex sul1-type integron, from the int gene to CR1 was amplified using PCR mapping and then sequenced. By cloning the region encompassing ampC and ampR, a recombinant plasmid (p413C) with an insert that conferred inducible resistance to ceftazidime was selected. The insert was found to contain blaDHA-1 and the regulatory gene ampR, which was downstream of blaDHA-1. This insert shared also part of pRBDHA's backbone carrying a complex integron (GenBank AJ971343). PCR and DNA sequencing results confirmed that the plasmid encoded at least three β-lactamase genes: blaTEM-1, blaSHV-12, and blaDHA-1, and a plasmid mediated resistance to quinolone (QnrB4).
DiscussionpAmpC have been found worldwide but are less common than ESBLs.3 They are emerging worldwide in various species of Enterobacteriaceae as a mechanism of acquired resistance to cefoxitin. In our study 1.6% (n=15) of the screened Enterobacteriaceae isolates were cefoxitin-resistant and produced plasmid-mediated AmpC β-lactamases. Prevalence of pAmpC in Algeria is not known, due to the limited number of epidemiological surveys. In Algeria, Iabaden et al. reported a prevalence of plasmid mediated AmpC β-lactamases of 2.18%.11 Mata et al. reported a significant increase in overall prevalence of Enterobacteriaceae carrying acquired AmpC in a Spanish hospital which was 0.43%, rising from 0.06% (1999) to 1.3% (2007).26 A prevalence of 12.5% was reported by Mohamudha et al. in India.27
Our study demonstrated that pAmpC-producing Enterobacteriaceae might be the cause of nosocomial and community infections in Algeria. Of note, we found that 40% of the cases were recovered from non-hospitalized patients. Isolation of pAmpC-producing Enterobacteriaceae from community was reported by many authors.28,29 Nursing homes and community-based sources of pAmpC-producers can pose a serious risk of transmission to hospitalized patients when infected or colonized patients are admitted. Gude et al. have found this resistance mechanism on isolates from community patients in a high rate, underscoring the need for close surveillance of these isolates.30 Several studies reported the isolation of pAmpC-producing Enterobacteriaceae isolates from food products, such as retail chicken meat, retail meat, and cheese.31–33 Thus, food chain might be a relevant vehicle for transmission of these enzymes in the community. They have also been detected in drinking water and river beaches.34 These sources could contribute to the spread of global pAmpC-producers in addition to a possible transmission of mobile genetic elements carrying resistance genes among strains.30
In Algeria, CMY-2 and DHA-1 were previously reported by Messai et al. (2006)10, Iabadene et al. (2009)11 and Nedjai et al. (2012).12 This is the first isolation of CMY-4 in clinical isolates (nosocomial and community infections) in Algeria. Thus, the first strain (K. pneumoniae 123) was isolated in 2005 from a patient hospitalized at Bejaia hospital (Algeria). The predominance of CMY-4 was consistent with worldwide observations. DHA-1 has been mostly reported in Asia.5,35
In our study, E. coli isolates were mainly groups B2 and D strains which are commonly extra-intestinal pathogenic strains, while phylogenetic groups A and B1 strains, usually commensal, were less frequent.36 CMY-2 production was reported in phylogenetic group D E. coli in humans and stray dogs.5,37
In the study of Mnif et al., the non-ST131-group B2 isolates, which were associated to CTX-M-15 ESBLs, had a higher frequency of several genes encoding key virulence factors such as adhesins hra, sfa/foc, papC and papG II, and the toxins hylA and cnf1 than had the ST131 isolates.38 In our study, a single isolate harbored several virulence genes iutA, papC and sfa/foc and belonged to phylogenetic group B2.
Our results showed that AmpC-producing K. pneumoniae isolates belonged to different sequence types. ST17 has been previously found in Cadiz, associated with CTX-M-15, in Freiburg and in Seoul, in Barcelona, associated with DHA-1.21,39–41 ST17 belongs to the ST17 complex, which contains four single-locus variants and six double-locus variants.41K. pneumoniae ST834 strains were previously involved in blaKPC dissemination in New Jersey.42 Besides the low number of isolates, we have detected two news sequences types: ST1617 and ST1618.
In this study, all isolates producing blaCMY-4 and blaDHA-1 co-expressed the broad-spectrum TEM-1 β-lactamase and three of them co-produced CTX-M and/or SHV ESBL. This enzyme combination complicates their detection and treatment. blaCMY-4 gene was located on broad host range A/C conjugative plasmid which was among the most commonly reported worldwide. In the last decades, Inc A/C plasmids have been associated with the spread of the AmpC beta lactamase CMY-2, in strains isolated from human, beef, chicken, turkey, and pork, revealing that this common plasmid backbone is broadly disseminated among resistant zoonotic pathogens.9,43
In our study, the genetic organization of blaCMY-4 and its variants was highly conserved. All the isolates carried the transposon-like element ISEcp1 (ISEcp1/ΔISEcp1-blaCMY-blc-sugE), as documented previously.44
The blaDHA-1gene was previously found on different plasmids of Inc groups A/C, FIA, FII, L/M, N, R and HI2 or of unknown Inc groups.9,11,26,41,45 Nevertheless, it is worth noting that blaDHA-1 gene was located on LVPK conjugative plasmid. Linkage of blaDHA-1 and qnrB4 genes of similar structures has been described in isolates of K. pneumoniae.8 The association among blaDHA-1, qnrB4, and aac(6′)-Ib-cr was reported before.46 The K. pneumoniae 413 strain in our study harbored a combination of β-lactamase genes (blaCTX-3, blaDHA-1, blaSHV-12 and blaTEM-1), PMQR determinants (qnrB4 gene) and aminoglycoside acetyltransferase gene (aac(6′)-Ib). Despite several investigations, we could not determine the origin of this multiresistant strain. To our knowledge, this is the first description of this association of genes including blaCTX-M-3 in the same strain. Identification of the sequences surrounding the blaDHA-1 gene found an ampR gene included in a complex sul-1-type integron that was likely similar to those previously reported.8
Use of antibiotics in both humans and animals, the global mobility of populations, and food products perpetuate the spread of multiresistant bacterial clones and resistance genes. Early identification of these organisms is necessary as the appropriate treatment might reduce the spread of these resistant strains and consequently mortality of hospitalized patients can be reduced. This emphasizes the need for such enzymes detection for preventing this emerging resistance into hospitals and for controlling its spread within the community. That will avoid therapeutic failures and nosocomial outbreaks.
Conflicts of interestThe authors declare no conflicts of interest.
We are grateful to J. Madoux for her contribution to this work. We thank the team of curators of the Institut Pasteur MLST databases for curating the data and making them publicly available at http://www.pasteur.fr/mlst.