To compare cultured microorganisms identified on endotracheal tubes biofilms through sonication technique with traditional tracheal aspirate collected at extubation of pediatric intensive care unit patients.
MethodsDemographic and epidemiological data were analyzed to identify factors possibly related with the microbiological profile of the two collection methods. Associations between categorical and continuous variables were analyzed using the chi-square or Fisher's exact test, or Student's t test. p-Value <0.05 were considered significant.
ResultsThirty endotracheal tubes and tracheal aspirates samples from 27 subjects were analyzed. Only one patient presented the clinical diagnosis of ventilator-associated pneumonia. Overall, 50% of bacteria were Gram-negative bacilli, followed by Gram-positive bacteria in 37%, and fungi in 10%. No statistically significant difference on the distribution of Gram-positive or Gram-negative bacteria (p=0.996), and fungi (p=0.985) were observed between the collection methods. Pseudomonas spp. was the most frequent microorganism identified (23.8%), followed by Streptococcus spp. (18.5%), Acinetobacter spp. (15.9%), coagulase-negative staphylococci (11.2%), and Klebsiella spp. (8.6%). Concordant results between methods amounted to 83.3%. Pseudomonas aeruginosa and Acinetobacter baumannii showed carbapenem resistance in 50% and 43.7% of the isolates, respectively. In general, cultures after endotracheal tubes sonication (non-centrifuged sonication fluid and centrifuged sonication fluid) yielded bacteria with higher rates of antimicrobial resistance compared to tracheal aspirates cultures. Additionally, in 12 subjects (40%), we observed discrepancies regarding microbiologic profiles of cultures performed using the collection methods.
ConclusionsOur study demonstrated that sonication technique can be applied to ET biofilms to identify microorganisms attached to their surface with a great variety of species identified. However, we did not find significant differences in comparison with the traditional tracheal aspirate culture approach.
Nosocomial pneumonia represents approximately one quarter of all nosocomial infections and tracheal intubation increases the risk for infection by six to 20 times higher.1,2 Among patients under mechanic ventilation on intensive care unit (ICU), ventilator-associated pneumonia (VAP) represents the most frequent type of nosocomial infection, which extends the length of hospitalization, increase mortality and costs of ICU treatment.1 The clinical diagnosis of VAP has been based upon local and systemic signs and symptoms of infection in addition to radiological criteria.1,2 Furthermore, bacteriological diagnosis using protected specimen brush (PSB), bronchoalveolar lavage (BAL), and protected endotracheal aspirates have been fully standardized but lack specificity, as it relies upon the identification of microorganisms growing in tracheal secretion.1–4 Nevertheless, bacteria growing in biofilms attached to the endotracheal tube may also play a role in the pathogenesis of VAP.5,6 Indeed, the knowledge of biofilm formation by microorganisms in a series of medical devices and its association to difficult-to-treat infections has become a concerning in the nosocomial setting.7 Therefore, the VAP represents a clear example of multifactorial infection in which the biofilm may be implicated not only on the pathogenesis, but equally on the high prevalence of multiresistant bacteria.8–12 The endotracheal tube (ET) allows direct entry of microorganisms from the oropharynx environment into the lower respiratory tract, whereas the innate immune system components are ineffective.12–14 Thus, ET allows microorganisms presenting natural ability to form biofilm to attach on the tube surface through production of a matrix of polysaccharides and proteins that provides protection and consequent survival in the environment.10 According to previous publications, bacteria within the biofilm can infect the lungs by several ways: through detachment of biofilm portions, thus reaching the lungs and by aspiration into deeper airways of aerosolized planktonic pathogens detached from the biofilm.13,14
Recently, microbial diagnosis of implant-associated infections has been incremented by using vortexing and sonication technique applied to dislodge sessile microorganisms attached to a variety of medical implants, by disrupting the polymer matrix on the surface of the biofilm while maintaining the integrity of the pathogen.15–17 Trampuz et al., Inacio et al., and Vandecandelaere et al. were some of the authors whose studies applied sonication for identifying pathogens from biofilms attached to different human implants.15,17–20 Despite the lack of data, ET sonication may be an interesting tool to increase the yield of detection of sessile pathogens and its possible effect on the pathogenesis of VAP.
Therefore, we aimed to compare the yield of cultured microorganisms collected by ET centrifuged and non-centrifuged sonication fluid with microorganisms collected by conventional endotracheal aspiration of intubated pediatric ICU patients.
Materials and methodsStudy populationWe performed a pilot microbiological cross-sectional study comparing the microbial colonization of ET submitted to sonication with the microbial yield of cultures of collected tracheal aspirates at the time of elective extubation. A total of 27 pediatric ICU patients under mechanical ventilation, from December 2012 to June 2014 at the Santa Casa de São Paulo School of Medicine (Brazil) were evaluated. The collection of tracheal aspirates was performed prior to extubation as part of the ICU physiotherapist routine in order to prevent bronchoaspirations. Subjects were excluded when clinical data were unavailable for analysis, when no tracheal aspirates were collected during extubation, or when contamination of ET occurred during extubation, transportation, or processing in the microbiology laboratory. As a cross-sectional study, study participants were not followed-up. The Research Ethics Committee at our institution approved the project and waiver of obtaining an informed consent, as the investigators have neither interfered in patient management nor in the indication of extubation.
Diagnosis of VAPThe diagnosis criteria for VAP were based on the CDC guidelines, in which patients on mechanical ventilation develop pneumonia 48h after intubation.21 The diagnostic triad consisted of clinical evidence of pulmonary infection, including fever, purulent secretions, leukocytosis or leukopenia, signs of respiratory distress and worsening gas exchange; radiologic suggestion of pulmonary infection, such as persistence of lung infiltrate, consolidations and cavitations; and microbial evidence of pulmonary infection.21
Collection of samples and sonication methodPrior to extubation, the ICU physiotherapy team routinely collects tracheal secretions to prevent bronchoaspiration in pediatric patients. This collected secretion, that otherwise would be discarded, was then used for the microbial analyses of the study. After extubation, the distal 10cm of the ET were chopped off and submitted to aseptic cleaning on its surface with either alcohol 70% or chlorhexidine. The cleaned portions of the ET were stored in a 50mL Falcon tubes with the addition of 20mL of sterile NaCl saline solution (0.9%). The samples were processed and cultivated within 6h from collection; on the laboratory the Falcon tubes were subjected to two cycles of vortexing for 30s using a Vortex-Genie 2 (Scientific Industries Inc., Bohemia, NY, USA), interspersed by sonication for 5min at a frequency of 40±2kHz with power density of 0.22W/cm2 ±0.04, using the ultrasonic washer model BactoSonic 14.2 (Bandelin Electronic GmbH & Co. KG, Germany), according to previous publications.15,17 After the second vortexing, aliquots of 0.1mL of non-centrifuged sonication fluid were inoculated on aerobic sheep blood and chocolate agar plates. The Falcon tube was then submitted to centrifugation for 5min at 2500rpm, the supernatant aspirated, leaving other aliquot of 0.1mL to be plated in the same media. Along the ET, tracheal secretion samples from the same patients were collected by the ICU team. The tracheal aspirate samples were also inoculated with a calibrated loop, using aliquots of 0.1mL onto blood and chocolate agar plates, and then interpreted after overnight incubation in appropriated conditions.
Microbiological analysisMicrobial analysis was performed for both centrifuged and non-centrifuged sonication fluid samples and for tracheal aspirates. The culture media were incubated for 48h, both at a temperature of 35–37°C. The chocolate agar was incubated in a capnophilic atmosphere. The plates showing microbial growth underwent quantification and identification of bacteria according to the routine established by the laboratory, considering the morphology and tinctorial properties displayed on Gram stain. In both methods, approximately 3–4 colony-forming units (CFU) were used as suspensions in saline until the inoculum reached 0.5 MacFarland scale (1.5×108CFU/mL). The inoculum was plated in Mueller Hinton Agar or Mueller Hinton Blood Agar. After 24-h incubation, the diameters of inhibition zones were measured and interpreted in accordance to standards for tested antimicrobial drugs.22 Catalase tests were performed on colonies of Gram-positive cocci for identifying Staphylococcus spp. and Streptococcus spp. DNAse tests differentiated Staphylococcus aureus from coagulase-negative staphylococci, while groups of Streptococcus spp. and Enterococcus spp. were identified by their hemolysis (alpha, beta or gamma), passing to hydrolysis of bilesculina tests and growth on 6.5% NaCl associated with susceptibility testing to optochin and bacitracin or CAMP Test, when necessary. The colonies of Gram-negative bacteria were identified by biochemical methods to determine their genus and species. The CLSI standardized disk diffusion method was used for all strains in the identification of susceptibility profile, and for vancomycin, we applied the Etest® (AB Biodisk, France) for determination of minimum inhibitory concentration (MIC).23 Antimicrobials used for testing the cultivated pathogens followed the recommendations of the CLSI.
Statistical dataSubject and epidemiological variables and other microbiological findings were summarized by using frequencies, percentages, means and standard deviations. Associations between categorical variables were analyzed using the chi-square or Fisher's exact test. Comparison of continuous variables was performed with the Student's t test. Differences were considered significant at a p-value <0.05.
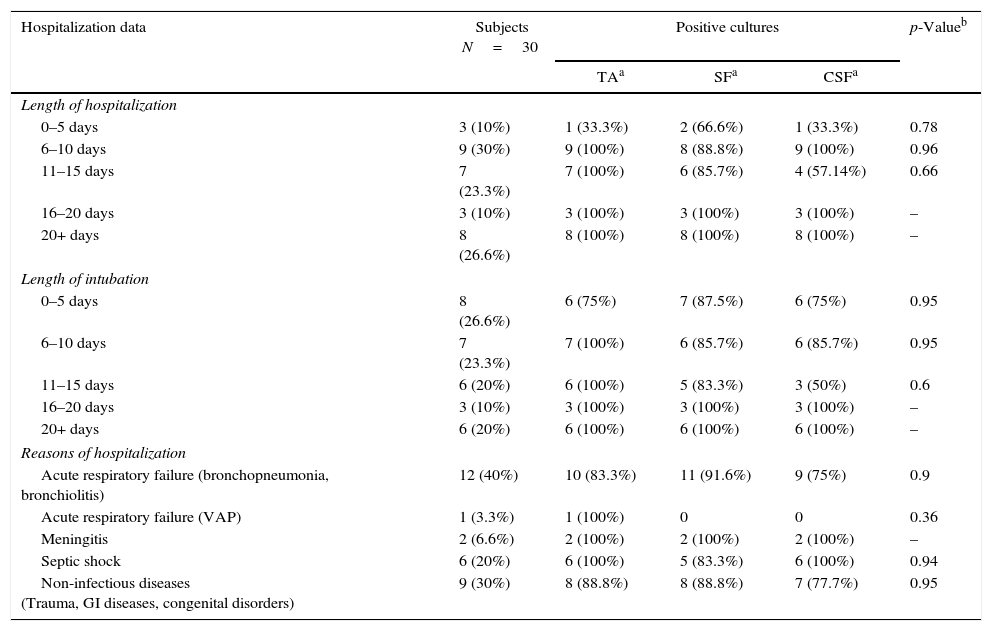
ResultsStudy population and empirical antimicrobial therapyThirty ET and tracheal aspirates samples were obtained from 27 patients. Three endotracheal tubes and TA samples were obtained from a single subject at different moments due to reintubation. The subjects mean age was 3.25±3.93 years, in which 17 patients (56.6%) were younger than one-year-old; 63.33% were male. Median length of hospital stay and time on mechanical ventilation were 16.2±12.76 days and 12.33±8.31 days, respectively. Only one subject presented the clinical diagnosis of VAP. Table 1 summarizes epidemiological data and clinical characteristics of the study population, such as length of hospital stay, days of mechanical ventilation, main reason for ICU admission, empirical antibiotic therapy. In addition, this table shows the frequencies of positive cultures according to the collection methods, namely tracheal aspirates (TA), sonication fluid (SF), and centrifuged sonication fluid cultures (CSF). There were no significant differences in the frequency of positive cultures according to the collection methods, after adjusting for epidemiological data, clinical characteristics, and type of empirical antibiotic therapy (Table 1).
Epidemiological data and prescribed antibiotics of study subjects and comparison with the frequency of positive cultures in each microbiological technique.
| Hospitalization data | Subjects N=30 | Positive cultures | p-Valueb | ||
|---|---|---|---|---|---|
| TAa | SFa | CSFa | |||
| Length of hospitalization | |||||
| 0–5 days | 3 (10%) | 1 (33.3%) | 2 (66.6%) | 1 (33.3%) | 0.78 |
| 6–10 days | 9 (30%) | 9 (100%) | 8 (88.8%) | 9 (100%) | 0.96 |
| 11–15 days | 7 (23.3%) | 7 (100%) | 6 (85.7%) | 4 (57.14%) | 0.66 |
| 16–20 days | 3 (10%) | 3 (100%) | 3 (100%) | 3 (100%) | – |
| 20+ days | 8 (26.6%) | 8 (100%) | 8 (100%) | 8 (100%) | – |
| Length of intubation | |||||
| 0–5 days | 8 (26.6%) | 6 (75%) | 7 (87.5%) | 6 (75%) | 0.95 |
| 6–10 days | 7 (23.3%) | 7 (100%) | 6 (85.7%) | 6 (85.7%) | 0.95 |
| 11–15 days | 6 (20%) | 6 (100%) | 5 (83.3%) | 3 (50%) | 0.6 |
| 16–20 days | 3 (10%) | 3 (100%) | 3 (100%) | 3 (100%) | – |
| 20+ days | 6 (20%) | 6 (100%) | 6 (100%) | 6 (100%) | – |
| Reasons of hospitalization | |||||
| Acute respiratory failure (bronchopneumonia, bronchiolitis) | 12 (40%) | 10 (83.3%) | 11 (91.6%) | 9 (75%) | 0.9 |
| Acute respiratory failure (VAP) | 1 (3.3%) | 1 (100%) | 0 | 0 | 0.36 |
| Meningitis | 2 (6.6%) | 2 (100%) | 2 (100%) | 2 (100%) | – |
| Septic shock | 6 (20%) | 6 (100%) | 5 (83.3%) | 6 (100%) | 0.94 |
| Non-infectious diseases (Trauma, GI diseases, congenital disorders) | 9 (30%) | 8 (88.8%) | 8 (88.8%) | 7 (77.7%) | 0.95 |
| Hospitalization data | Subjects N=27 | Positive cultures | p-Valueb | ||
|---|---|---|---|---|---|
| TAa | SFa | CSFa | |||
| Antibiotics prescribed | |||||
| Ceftazidime | 1 (3.70%) | 1 (100%) | 1 (100%) | 1 (100%) | – |
| Amikacin | 4 (14.81%) | 4 (100%) | 4 (100%) | 4 (100%) | – |
| Metronidazole | 3 (11.11%) | 3 (100%) | 2 (66.6%) | 3 (100%) | 0.88 |
| Ceftriaxone | 15 (55.55%) | 14 (93.3%) | 14 (93.3%) | 12 (80%) | 0.90 |
| Clindamycin | 4 (14.81%) | 4 (100%) | 3 (75%) | 4 (100%) | 0.91 |
| Vancomycin | 15 (55.55%) | 15 (100%) | 14 (93.3%) | 13 (86.6%) | 0.93 |
| Meropenem | 15 (55.55%) | 15 (100%) | 14 (93.3%) | 13 (86.6%) | 0.93 |
| Fluconazole | 8 (29.62%) | 8 (100%) | 7 (87.5%) | 7 (87.5%) | 0.95 |
| Cefazolin | 2 (7.40%) | 1 (50%) | 1 (50%) | 1 (50%) | – |
| Micafungin | 2 (7.40%) | 2 (100%) | 2 (100%) | 2 (100%) | – |
| Cefepime | 3 (11.11%) | 3 (100%) | 2 (66.6%) | 3 (100%) | 0.88 |
| Oxacillin | 13 (48.14%) | 12 (92.3%) | 11 (84.6%) | 10 (77%) | 0.91 |
| Piperacillin/tazobactam | 2 (7.40%) | 2 (100%) | 2 (100%) | 2 (100%) | – |
| Crystalline penicillin | 3 (11.11%) | 3 (100%) | 3 (100%) | 3 (100%) | – |
| Clarithromycin | 2 (7.40%) | 2 (100%) | 2 (100%) | 2 (100%) | – |
| Amphotericin B | 1 (3.70%) | 1 (100%) | 1 (100%) | 1 (100%) | – |
| Ampicillin/sulbactam | 1 (3.70%) | 1 (100%) | 1 (100%) | 1 (100%) | – |
| Azithromycin | 1 (3.70%) | 1 (100%) | 1 (100%) | 1 (100%) | – |
TA, tracheal aspirate; SC, non-centrifuged sonication fluid; CSF, centrifuged sonication fluid.
Hospitalization data were summarized as frequencies and percentages or median and compared using the chi-square or Fisher's exact test as appropriate to categorical variables and t-test as appropriate to continuous variables. All tests were two sided and p-values less than 0.05 were considered statistically significant.
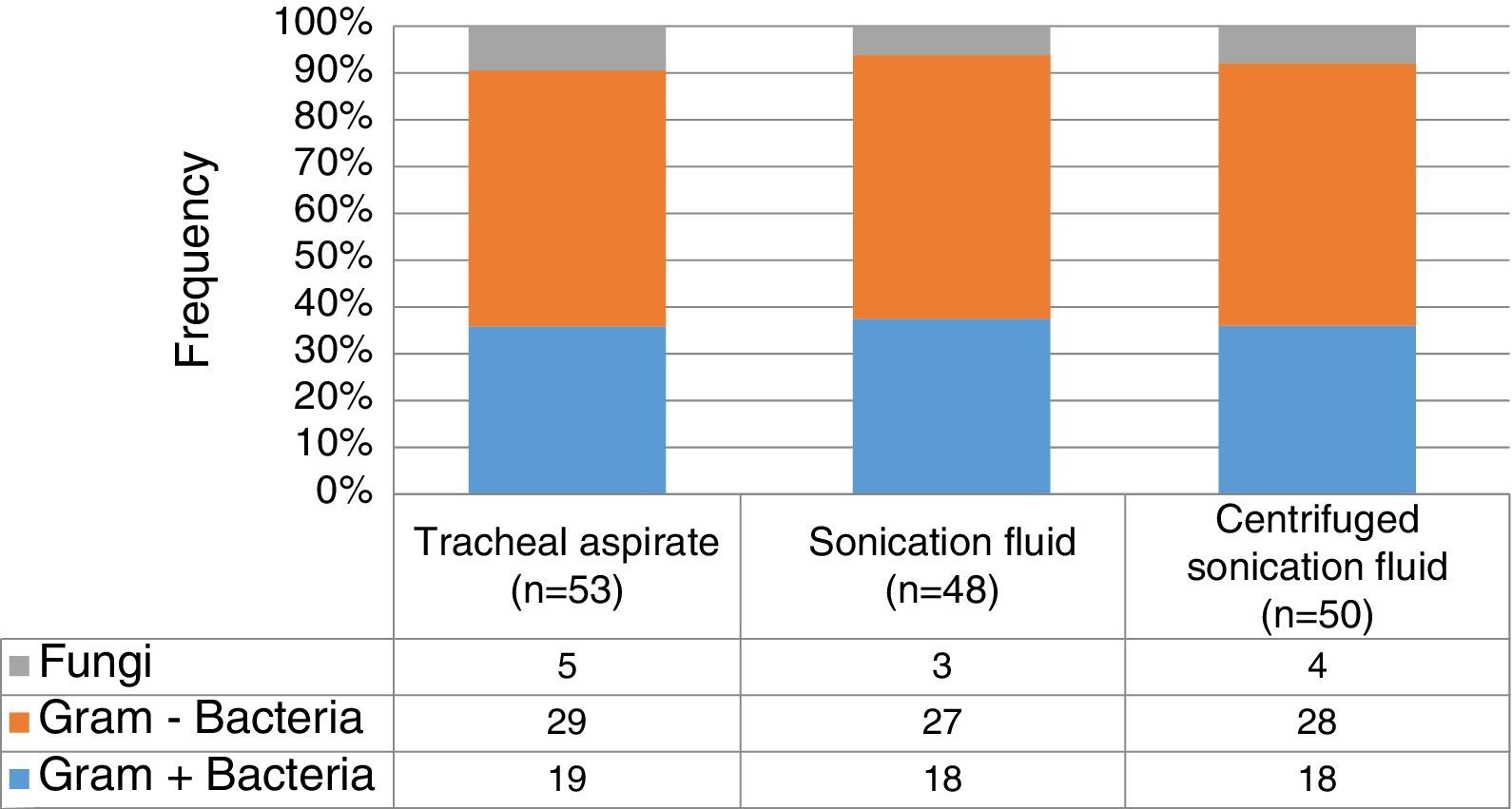
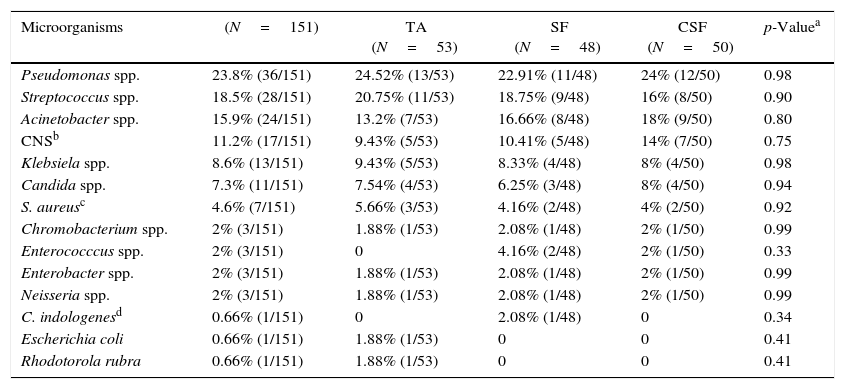
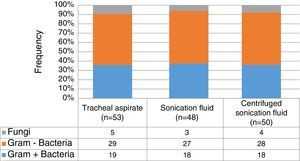
In general, up to 50% of identified microorganisms were Gram-negative bacilli, followed by Gram-positive bacteria in 37%, whereas fungi represented less than 10% (Fig. 1). No statistically significant differences on the distribution of Gram-positive or negative bacteria (p=0.99), and fungi (p=0.98) according to the collection three methods. Pseudomonas spp. (23.8%) and Streptococcus spp. (18.5%) were the most frequently identified pathogens, followed by Acinetobacter spp. (15.9%), coagulase-negative staphylococci (11.2%), Klebsiella spp. (8.6%) and Staphylococcus aureus (4.6%). Interestingly, there was no statistically significant difference on the distribution of microorganisms isolated from sonication fluid (centrifuged and non-centrifuged) compared with tracheal aspirate. Table 2 summarizes the frequencies of microorganisms cultured according to the collection method ET submitted to sonication and the method of conventional tracheal aspirate.
Distribution of species of bacteria and fungi according to its frequencies in cultures of tracheal aspirates, sonication fluid, and centrifuged sonication fluid.
| Microorganisms | (N=151) | TA (N=53) | SF (N=48) | CSF (N=50) | p-Valuea |
|---|---|---|---|---|---|
| Pseudomonas spp. | 23.8% (36/151) | 24.52% (13/53) | 22.91% (11/48) | 24% (12/50) | 0.98 |
| Streptococcus spp. | 18.5% (28/151) | 20.75% (11/53) | 18.75% (9/48) | 16% (8/50) | 0.90 |
| Acinetobacter spp. | 15.9% (24/151) | 13.2% (7/53) | 16.66% (8/48) | 18% (9/50) | 0.80 |
| CNSb | 11.2% (17/151) | 9.43% (5/53) | 10.41% (5/48) | 14% (7/50) | 0.75 |
| Klebsiela spp. | 8.6% (13/151) | 9.43% (5/53) | 8.33% (4/48) | 8% (4/50) | 0.98 |
| Candida spp. | 7.3% (11/151) | 7.54% (4/53) | 6.25% (3/48) | 8% (4/50) | 0.94 |
| S. aureusc | 4.6% (7/151) | 5.66% (3/53) | 4.16% (2/48) | 4% (2/50) | 0.92 |
| Chromobacterium spp. | 2% (3/151) | 1.88% (1/53) | 2.08% (1/48) | 2% (1/50) | 0.99 |
| Enterococccus spp. | 2% (3/151) | 0 | 4.16% (2/48) | 2% (1/50) | 0.33 |
| Enterobacter spp. | 2% (3/151) | 1.88% (1/53) | 2.08% (1/48) | 2% (1/50) | 0.99 |
| Neisseria spp. | 2% (3/151) | 1.88% (1/53) | 2.08% (1/48) | 2% (1/50) | 0.99 |
| C. indologenesd | 0.66% (1/151) | 0 | 2.08% (1/48) | 0 | 0.34 |
| Escherichia coli | 0.66% (1/151) | 1.88% (1/53) | 0 | 0 | 0.41 |
| Rhodotorola rubra | 0.66% (1/151) | 1.88% (1/53) | 0 | 0 | 0.41 |
TA, tracheal aspirate; SC, non-centrifuged sonication fluid; CSF, centrifuged sonication fluid.
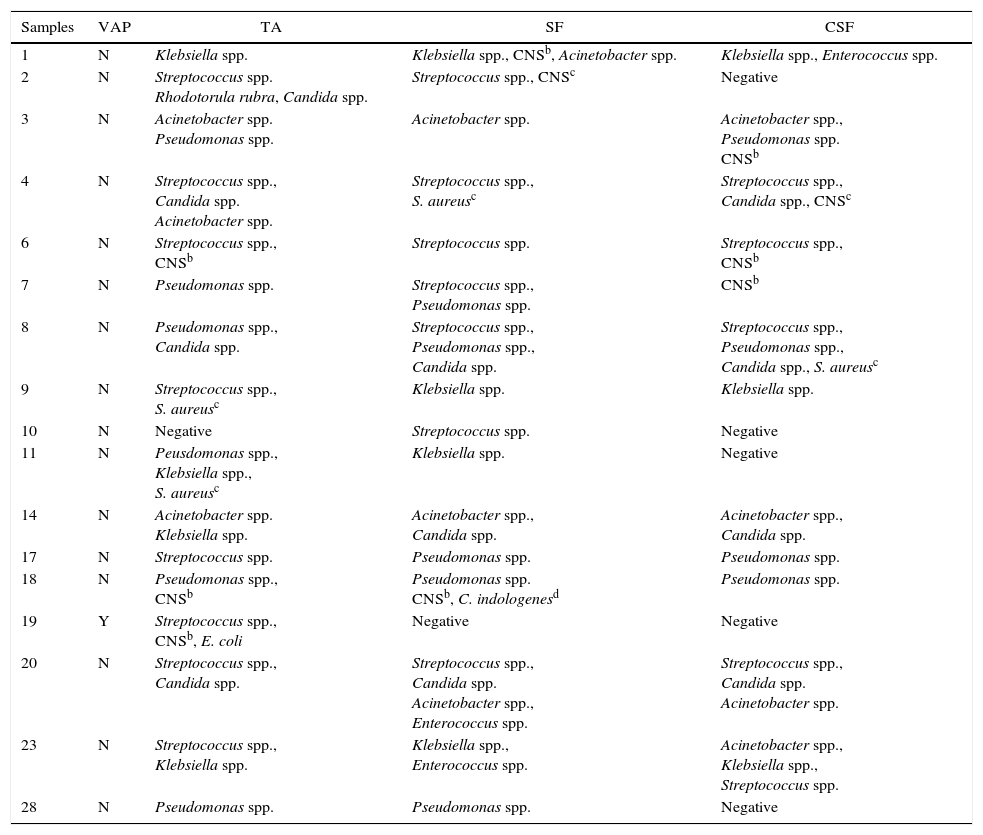
In all but one ET and TA samples no microbial growth was detected neither on SF, CSF nor on TA cultures (3.3%, 1/30). The global concordance (positive and negative results) between TA, SF, and CSF was 83.3%. There were five discordant results, in which only one subject had sonication fluid culture-positive and TA culture-negative result. On the other hand, the only subject showing clinical diagnosis of VAP had TA culture-positive (Escherichia coli) and SF/CSF culture-negative results. Surprisingly, subject #11 was not clinically diagnosed with VAP, but TA cultures yielded Pseudomonas spp., Klebsiella spp., S. aureus, and only Klebsiella spp. on SF cultures. Two subjects presented CSF culture-negative results, but cultures of SF and TA were positive and discrepancy was detected in one of them (Table 3). Additionally, we found microbiological discrepancies (positive cultures on TA, SF, and CSF, but different microbiological profiles) among 12 (40%) subjects. Table 3 summarizes the discrepant and discordant results between TA, SF, and CSF cultures.
Subjects with discordant and discrepanta culture results of tracheal aspirates, sonication fluid, and centrifuged sonication fluid.
| Samples | VAP | TA | SF | CSF |
|---|---|---|---|---|
| 1 | N | Klebsiella spp. | Klebsiella spp., CNSb, Acinetobacter spp. | Klebsiella spp., Enterococcus spp. |
| 2 | N | Streptococcus spp. Rhodotorula rubra, Candida spp. | Streptococcus spp., CNSc | Negative |
| 3 | N | Acinetobacter spp. Pseudomonas spp. | Acinetobacter spp. | Acinetobacter spp., Pseudomonas spp. CNSb |
| 4 | N | Streptococcus spp., Candida spp. Acinetobacter spp. | Streptococcus spp., S. aureusc | Streptococcus spp., Candida spp., CNSc |
| 6 | N | Streptococcus spp., CNSb | Streptococcus spp. | Streptococcus spp., CNSb |
| 7 | N | Pseudomonas spp. | Streptococcus spp., Pseudomonas spp. | CNSb |
| 8 | N | Pseudomonas spp., Candida spp. | Streptococcus spp., Pseudomonas spp., Candida spp. | Streptococcus spp., Pseudomonas spp., Candida spp., S. aureusc |
| 9 | N | Streptococcus spp., S. aureusc | Klebsiella spp. | Klebsiella spp. |
| 10 | N | Negative | Streptococcus spp. | Negative |
| 11 | N | Peusdomonas spp., Klebsiella spp., S. aureusc | Klebsiella spp. | Negative |
| 14 | N | Acinetobacter spp. Klebsiella spp. | Acinetobacter spp., Candida spp. | Acinetobacter spp., Candida spp. |
| 17 | N | Streptococcus spp. | Pseudomonas spp. | Pseudomonas spp. |
| 18 | N | Pseudomonas spp., CNSb | Pseudomonas spp. CNSb, C. indologenesd | Pseudomonas spp. |
| 19 | Y | Streptococcus spp., CNSb, E. coli | Negative | Negative |
| 20 | N | Streptococcus spp., Candida spp. | Streptococcus spp., Candida spp. Acinetobacter spp., Enterococcus spp. | Streptococcus spp., Candida spp. Acinetobacter spp. |
| 23 | N | Streptococcus spp., Klebsiella spp. | Klebsiella spp., Enterococcus spp. | Acinetobacter spp., Klebsiella spp., Streptococcus spp. |
| 28 | N | Pseudomonas spp. | Pseudomonas spp. | Negative |
VAP, ventilator-associated pneumonia; TA, tracheal aspirate; SC, non-centrifuged sonication fluid; CSF, centrifuged sonication fluid.
Regarding antimicrobial resistance, there were different patterns of susceptibility to antimicrobials among the same microorganisms identified on TA, SF, and SCF cultures. Of interest, Pseudomonas aeruginosa and Acinetobacter baumannii showed carbapenem resistance in 50% and 43.7% of the isolates, respectively. In general, cultures after ET sonication (SF and CSF) yielded bacteria with higher rates of antimicrobial resistance compared to TA cultures. For example, carbapenem-resistant and cefepime-resistant P. aeruginosa isolated from SF, CSF, and TA cultures were 35%, 35%, 29%, and 37.5%, 37.5%, 25%, respectively. Carbapenem-resistant A. baumannii isolated from SF, CSF, and TA cultures were 28.5%, 42.8%, 28.5%, respectively. Methicillin-resistant S. aureus (MRSA) was identified in 28.5% of the isolates, and Klebsiella pneumoniae Carbapenemase (KPC) was isolated in only one subject, identified in TA, SF, and SCF cultures.
DiscussionMicrobiological data showing ET biofilm flora and upper airway colonization remains insufficient, even though it has been frequently recognized as potential risk factors for nosocomial pneumonia. Therefore, we aimed to provide information regarding the microbiota attached to ET inner surfaces biofilms by use of sonication and compared to conventional tracheal aspirates cultures, among colonized pediatric ICU patients. Sonication has been frequently applied for dislodging biofilm sessile pathogens, and therefore validated as an adds-on technique for microbial diagnosis of different human device-associated infection.15–17,19 Vandecandelaere et al. applied sonication on ET for biofilm detachment and compared with TA surveillance cultures among mechanical ventilated adult patients, and found correlation between antibiotic-resistant pathogens identified either on ET biofilms and tracheal secretion cultures.20 Gil-Perotin et al. analyzed the formation of ET biofilm and its implication to VAP by scraping the biofilm from the inner surface of ET followed by vortexing. Biofilm was found almost universally on ET and 50% of bacteria associated to VAP were also cultured from biofilms.11
To our knowledge, no previous studies have applied sonication and vortexing on ET for biofilm microbial analysis in the ICU pediatric population. Unlike others authors, we investigated microbial biofilm formation on ET mainly among subjects with no clinical diagnosis of VAP (with one exception), even though they were all on systemic antimicrobial therapy for varied reasons. We found that even on colonized mechanically ventilated pediatric patients, there were a high prevalence (96.7%) of ET biofilm formation after intubation. Previous authors have already acknowledged the association between upper respiratory tract colonization, microaspiration, and biofilm formation on the surface of ET as risk factor for nosocomial pneumonia.5,6,8,9,11 Another important aspect of our study was the special attention to investigate the microbial results of SF using a concentration step. Indeed, many authors have added a SF concentration step using samples centrifugation to identify higher number of microorganisms on the sample.15–19,24 At least for biofilms attached to ET inner surface, centrifugation of sonication fluid was not statistically superior to non-centrifuged SF to increase microbial diagnosis.
Along with other results,5,11 in this study TA and SF cultures yielded mostly Gram-negative bacilli, mainly P. aeruginosa and A. baumannii (40%). These pathogens have been highly associated with severe acute and chronic nosocomial respiratory infections as they are easily capable of expressing biofilm-forming and antibiotic-resistant genes.1,5–11,20,25,26 Previous studies analyzed the microorganisms present in dental plaque of ICU patients and showed that more isolates of P. aeruginosa were identified when compared to the general population.27–28 Besides, Pseudomonas spp., although not a common inhabitant of oral microflora is an important respiratory pathogen frequently associated with late-onset VAP.7,11,27 The remarkable ability of P. aeruginosa to adhere and form complexes biofilm structures in sputum, lung tissues, and ET and therefore producing difficult-to-treat chronic infections, such as cystic fibrosis and VAP, was the very beginning of studies connecting biofilm science with medicine.5,13,14 Broad-spectrum antimicrobial therapy was offered for all patients in our studied sample, which might have contributed to the high prevalence of A. baumannii isolated. Indeed, late-onset VAP have been frequently caused by multidrug-resistant A. baumannii, an environmental opportunistic biofilm-forming low-virulent pathogen capable of surviving to moist and dry conditions for a prolonged period, and associated to previous use of systemic empirical antibiotics.25,26,29,30 Of note, common oral microbiota such as Streptococcus spp., which presents a wide variety of species, including S. mutans, a biofilm promoter, were also frequently isolated.27 Coagulase-negative staphylococci, which have a well-known genetic predisposition to form biofilms, were also frequently detected on cultures. Our results seemed to be in accordance with a recent study that applied traditional culture methods and molecular techniques to assess the microbial diversity of ET biofilms, and eventually identified coagulase-negative staphylococci, mainly S. epidermidis as the most frequently encountered organism on cultures.18
In our study, the overall microbial culture results observed from each collection technique were concordant (83%), which suggests that culture of all three techniques (TA, SF, and CSF) may have isolated both sessile and planktonic cells. A study by Vandecandelaere et al. also suggests an agreement of results between surveillance cultures and ET biofilm cultures using sonication technique.20 However, in a situation of VAP, according to ESCMID guideline for the diagnosis and treatment of biofilm infections, it is difficult to determine if the biofilm within the ET can be either the primary source of infection or merely a concomitant colonized site.6 Also, respiratory secretions can accumulate in the ET leading to a vast concentration of planktonic pathogens in airway secretion, underestimating the presence of sessile pathogens.
Regarding the presence of discordant results among reintubated patients and discrepancy in antibiotics susceptibility, biological variation and change of microbial flora due to a longer stay in ICU might be the reason behind the discordance, once common in-hospital pathogens were found at the second moment, like Acinetobacter spp., P. aeruginosa and Candida albicans. We also hypothesize that planktonic forms are more susceptible to create drug-resistant strains and are easily detectable through aspirate cultures. Besides, the biofilm itself represents a resistance mechanism to antibiotics, and once the bacteria are dispersed they are detected and rapidly become susceptible to antibiotics. As no significant differences were found between the techniques assessed in this study, the tracheal aspirate could be an option for the identification of pathogens from the biofilm, in which was found a similar microbial sessile flora when compared to the isolates of planktonic pathogens. This study, however, had a small sample size, which limited the microbiological findings, and the strength of our statistical analysis. The absence of more patients diagnosed with VAP could also underestimate the findings by showing less pathogens, as different sensibility patterns in comparison to the results obtained. Although our study demonstrated 83.3% of concordance between techniques we do not recommend using sonication as a diagnostic test without further studies, which should be performed to better understand the relation between VAP pathogenesis and ET biofilms.
Conflicts of interestThe authors declare no conflicts of interest.
We thank Eliete Celestino, Lucia Hiromi Kawai, and Maria Aparecida Murça for their outstanding technical assistance and Regina Grigolli Cesar for her support in the collection of tracheal aspirate among the ICU pediatric subjects. This work was supported by FAPESP (Fundação de Amparo a Pesquisa do Estado de São Paulo) under grant numbers 2013/00129-3 and 00130-1. The sponsor had no involvement in the study design, in the collection, analysis, and interpretation of data, in the writing of the manuscript, or in the decision to submit the manuscript for publication.