To evaluate the prevalence of some virulence genes among 510 clinical Enterococcus spp. isolates and to assess the association of those genes with the species, infection site, and patient group (inpatients/outpatients).
MethodsAdhesins genes (aggregation substances agg and asa1 of Enterococcus faecalis and Enterococcus faecium, respectively), enterococcal surface protein (esp), endocarditis-specific antigen A (efaA), collagen-binding proteins (ace/acm)); invasins (hyaluronidase (hyl) and gelatinase (gelE)); cytotoxines (activation of cytolysin (cylA) in E. faecalis); and modulators of the host immunity and inflammation (enhanced expression pheromone (eep) in E. faecalis) were detected by polymerase chain reaction.
ResultsThe overall prevalence was: esp – 44.3%, agg/asa1 – 38.4%, ace/acm – 64.3%, efaA – 85.9%, eep – 69.4%, gelE – 64.3%, hyl – 25.1%, and cylA – 47.1%. E. faecalis isolates had significantly higher frequency of adhesin genes (esp and agg/asa1) and gelatinase in comparison to E. faecium. Multiple virulence genes in E. faecalis were significantly more prevalent than in E. faecium isolates. Domination of E. faecium with or without only one gene compared to the isolates of E. faecalis were found. Enterococcus spp. isolates obtained from outpatients compared to inpatients isolates had significantly higher frequency of agg/asa1, eep, gelE and cylA. Some adhesins genes (esp, agg/asa1 and efaA) had higher prevalence among the non-invasive Enterococcus spp. isolates compared to those causing invasive bacteremia, while ace/acm revealed higher dissemination in isolates causing invasive infections compared to non-invasive isolates.
ConclusionMost E. faecalis attaches to abiotic surfaces in hospital environment, which correlates with higher prevalence of gene encoding for virulence factors involved in biofilm formation, such as enterococcal surface protein, aggregation substance, and gelatinase. The intestinal tract is an important reservoir for opportunistic enterococcal pathogens and allows them to access infectious sites through different virulence factors, demonstrated in outpatient isolates in this study.
Enterococci (mostly Enterococcus faecalis and Enterococcus faecium) are natural inhabitants of the gastrointestinal tract of humans and animals,1 but are also found in other anatomical sites, including the vagina and oral cavity,2 and in the soil, water, plants, and food.3–5 Several reports have documented that the two most important species are among the leading causes of opportunistic human infections,6 including urinary tract infections,7 infections of the surgical site and burn wound infections,8,9 bacteremia and sepsis,10 endocarditis,11 cholecystitis,12 peritonitis,13 neonatal meningitis,14 and others.
The severe infections caused by Enterococcus spp. are difficult to treat due to the organism's capacity to survive in the hospital environment, their intrinsic resistance to many antimicrobials, their remarkable ability to acquire further resistance mechanisms toward strategic antibiotics during the treatment period and a variety of virulence factors.15
Virulence factors contribute to the pathogenesis of enterococcal infections through mediation of adhesion, colonization and invasion into the host tissues, modulation of the host immunity, and extracellular production of enzymes and toxins, which enhance the severity of the infection.16 Many virulence factors, such as the enterococcal surface protein (Esp), aggregation substance, capsule formation and gelatinase, are involved in bacterial adherence to host cells and/or in biofilm formation on abiotic surfaces in hospital environment.17–21 Biofilm production has an important role in the pathogenesis of enterococcal infections and also favors disease sustenance because of restricted penetration of antimicrobials.19 Invasion is usually facilitated by damage to host tissues and presence of virulence factors, including hyaluronidase and gelatinase, which assist in the advancement and further survival in newly infected places.21
Currently, our knowledge about the possible relationship between the presence of virulence factors and their implication in human enterococcal infections is still limited.
The aim of the present study was to examine the prevalence of genes encoding cell-associated and extracellular virulence factors in clinically relevant isolates of Enterococcus spp. and to assess the association of those genes with species, infection site, and patient population (inpatients or outpatients).
Materials and methodsBacterial isolatesA collection of 510 non-duplicate clinical strains of Enterococcus spp., causing symptomatic infections, was investigated. All urinary tract infections were associated with significant bacteriuria (≥105CFU/mL). The isolates were recovered between June 2013 and June 2015 from 398 inpatients and 112 outpatients in seven large Bulgarian hospitals. They were obtained from urine (n=256), surgical wound or abscesses (n=138), genital tract samples (n=74), blood (n=26), peritoneal fluid (n=6), lower respiratory tract samples (n=8), and bile (n=2). E. faecalis ATCC 29212 and E. faecium ATCC 19434 were used as control strains for species identification with biochemical and molecular genetic techniques.
Culture mediaHiCrome E. faecium Agar Base (Himedia Labs) was applied to isolate Enterococcus spp. strains. E. faecalis forms blue colonies, while E. faecium gives green colonies, surrounded by yellowish coloring of the ambient.
Biochemical identification with commercial kits and systemsSpecies identification was done using API Rapid ID 32 Strep (bioMérieux), BBL Crystal Gram-positive ID kit (Becton Dickinson) and the automated system VITEK 2 (bioMérieux).
DNA isolationTotal DNA from all used strains was extracted with GenElute™ Bacterial Genomic DNA Kit (Sigma–Aldrich), according to the manufacturer's instructions, from 3mL overnight cultures inoculated with a single colony.
Molecular genetic genus and species identificationPolymerase chain reaction (PCR) amplification of the 16S rRNA gene of Enterococcus spp. was used for genus identification of all isolates included in this study. The sodA genes encoding the enzyme manganese-dependent superoxide dismutase in the most common enterococci (E. faecalis, E. faecium and Enterococcus durans) were detected with multiplex PCR. E. faecalis identification was confirmed by PCR for the eda1 gene (encoding 2-keto-3-deoxy-6-phosphogluconate aldolase, which is an enzyme involved in the Entner–Doudoroff pathway and is species-specific for E. faecalis). Oligonucleotides used as primers and amplification conditions were previously described.22
PCR assay for detection of virulence genesPCR amplification was performed in order to confirm the presence of genes coding for different virulence factors: adhesins (aggregation substance – agg and asa1 of E. faecalis and E. faecium, respectively), enterococcal surface protein (esp for both species), endocarditis-specific antigen A (efaA for both species), collagen-binding proteins (ace/acm); invasins (extracellular metalloendopeptidase also called gelatinase – gelE for both species, hyaluronidase – hyl for both species); cytotoxines (activation of cytolysin (class I. bacteriocin) – cylA in E. faecalis) and modulators of the host immunity and inflammation (sex pheromones – enhanced expression pheromone, eep in E. faecalis). Oligonucleotides used for specific amplification primers (Alpha DNA) were previously published in the literature23–29 and are shown in eTable 1 (see the E-supplemental material).
Supplementary Table 1 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bjid.2015.11.011.
PCR was carried out with 2μL template DNA, 0.25μM of each primer, 0.2mM deoxyribonucleoside triphosphates, 1× reaction buffer, 2mM MgCl2 and 0.5U Prime Taq DNA polymerase (Genet Bio) in a total volume of 25μL. The DNA was amplified in Gene Pro Thermal Cycler (Bioer) using the following protocol: initial denaturation (94°C for 5min), followed by 25–30 cycles of denaturation (94°C for 35–45s), annealing (50–68°C, from 45s to 1min) and extension (72°C, from 45s to 1min 35s), with a single final extension of 7min at 72°C. PCR products were separated in 1% agarose gel for 50–110min at 130V, stained with ethidium bromide (0.5μg/mL) and detected by UV transillumination (wavelength 312nm). Amplified genes were identified on the basis of fragment size presented in eTable 1.
Statistical analysisThe distribution of virulence genes with respect to both species and isolate origin was compared using Student's t-test. For a simple comparison tests a p-value below 0.05 was considered statistically significant. In order to counteract the problem of multiple comparisons, when they were used, a Bonferroni correction with a critical value of p=0.00625 was applied.
ResultsPrevalence of virulence genes among all studied Enterococcus spp. isolates (n=510)The established overall frequencies of the studied genes encoding adhesins (esp, agg/asa1, ace/acm, efaA), modulators of the host immunity and inflammation (eep), invasins (gelE, hyl), and cytotoxins (cylA) were as it follows: 44.3%, 38.4%, 64.3%, 85.9%, 69.4%, 64.3%, 25.1%, and 47.1%.
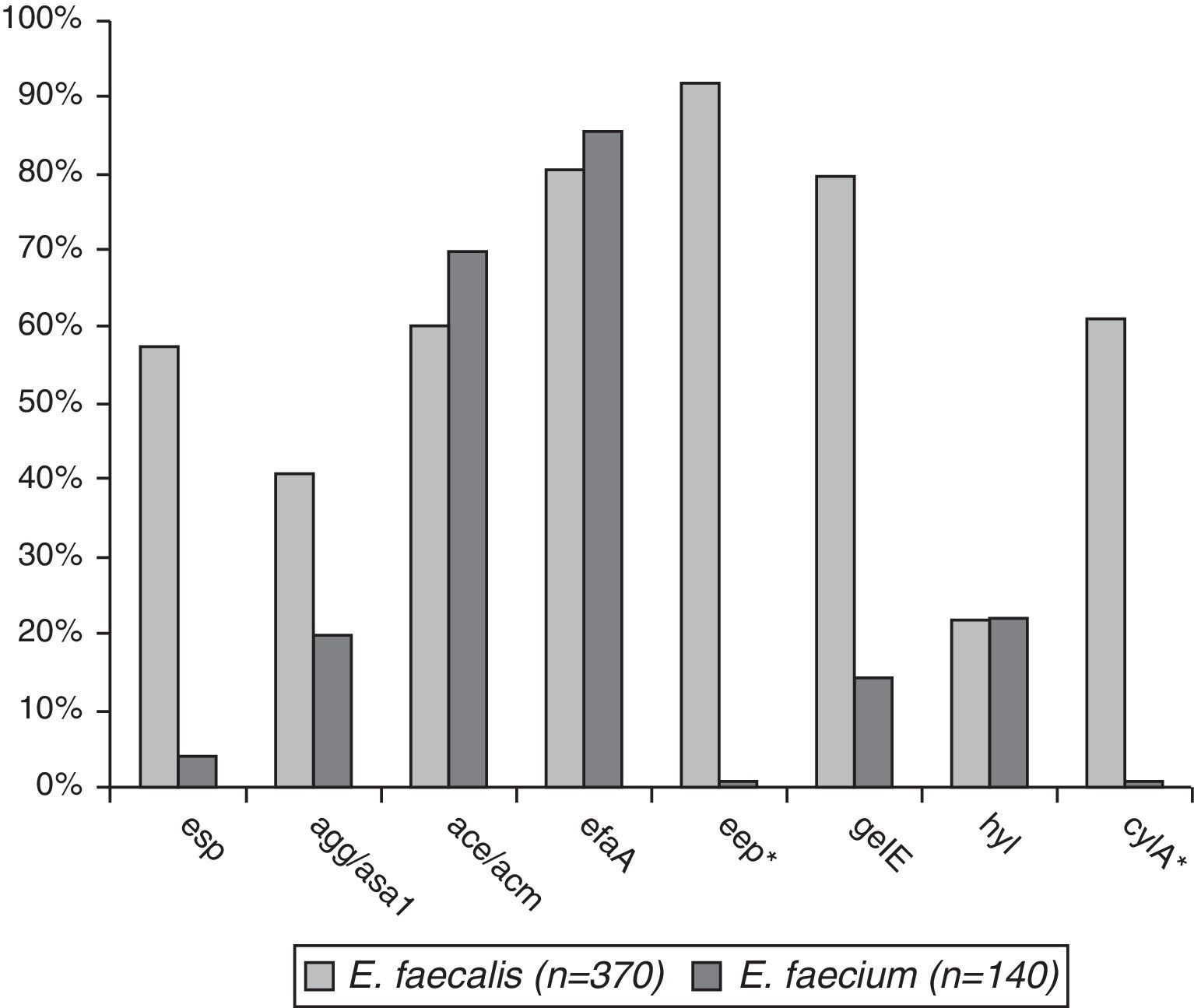
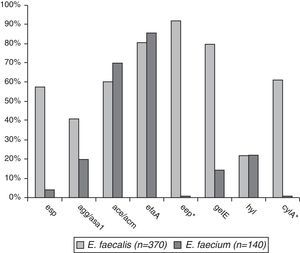
Comparative prevalence of virulence genes among E. faecalis (n=370) and E. faecium (n=110)The prevalence of virulence genes in the two species was esp – 59.5% in E. faecalis and 4.3% in E. faecium (p<0.001), agg/asa1 – 44.3% vs. 22.8% (p<0.001), ace/acm – 61.1%/72.8%, efaA – 84.9%/88.5%, gelE – 82.2%/17.1% (p<0.001), and hyl – 25.3%/24.2%. eep was found in 95.7% of E. faecalis isolates and cylA in 64.9% (Fig. 1).
The genes associated with virulence potential of the studied E. faecalis and E. faecium isolates are shown in Tables 1A and 1B. It should be pointed out in these tables the significantly higher prevalence of E. faecalis with multiple virulence determinants (over 4) compared to E. faecium (71.9% vs. 1.5%, p<0.001), whereas isolates with only one determinant significantly prevailed in E. faecium compared to E. faecalis (50.0%/1.1%, p<0.001).
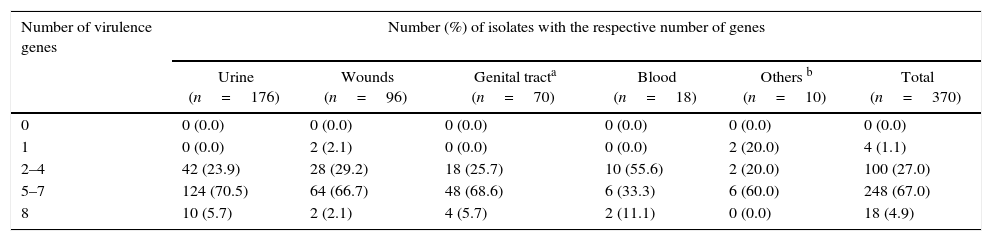
Virulence potential (depending the number of present virulence factor genes) in 370 E. faecalis isolates from different infection sites.
| Number of virulence genes | Number (%) of isolates with the respective number of genes | |||||
|---|---|---|---|---|---|---|
| Urine (n=176) | Wounds (n=96) | Genital tracta (n=70) | Blood (n=18) | Others b (n=10) | Total (n=370) | |
| 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 1 | 0 (0.0) | 2 (2.1) | 0 (0.0) | 0 (0.0) | 2 (20.0) | 4 (1.1) |
| 2–4 | 42 (23.9) | 28 (29.2) | 18 (25.7) | 10 (55.6) | 2 (20.0) | 100 (27.0) |
| 5–7 | 124 (70.5) | 64 (66.7) | 48 (68.6) | 6 (33.3) | 6 (60.0) | 248 (67.0) |
| 8 | 10 (5.7) | 2 (2.1) | 4 (5.7) | 2 (11.1) | 0 (0.0) | 18 (4.9) |
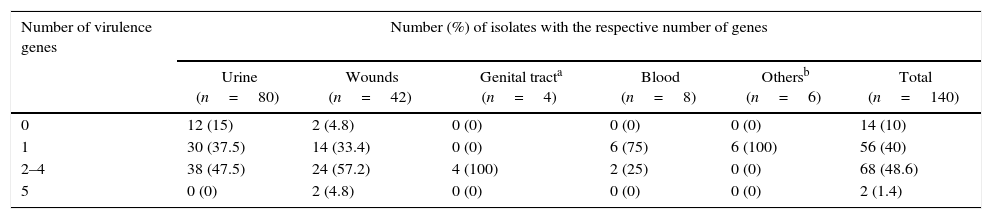
Virulence potential (depending the number of present virulence factor genes) in 140 E. faecium isolates from different infection sites.
| Number of virulence genes | Number (%) of isolates with the respective number of genes | |||||
|---|---|---|---|---|---|---|
| Urine (n=80) | Wounds (n=42) | Genital tracta (n=4) | Blood (n=8) | Othersb (n=6) | Total (n=140) | |
| 0 | 12 (15) | 2 (4.8) | 0 (0) | 0 (0) | 0 (0) | 14 (10) |
| 1 | 30 (37.5) | 14 (33.4) | 0 (0) | 6 (75) | 6 (100) | 56 (40) |
| 2–4 | 38 (47.5) | 24 (57.2) | 4 (100) | 2 (25) | 0 (0) | 68 (48.6) |
| 5 | 0 (0) | 2 (4.8) | 0 (0) | 0 (0) | 0 (0) | 2 (1.4) |
Table 2 lists the prevalence of virulence genes of all enterococci isolates, as well as the prevalence in each enterococcal species, broken down by the infection site. The following are the most important results found:
- •
The genes for some adhesins (esp; agg/asa1 and efaA) were significantly more prevalent among the non-invasive isolates of Enterococcus spp. (51.4% in genital tract infection (GTI) isolates, 39.1–59.5% in wound and GTI isolates, and 93.8% in urinary tract infection (UTI) isolates) compared to the invasive bacteremia isolates (respectively, 23.1%, 15.4%, and 61.5%) (p<0.001–0.05), while ace/acm revealed a significantly higher frequency (p<0.02–0.05) in invasive isolates (84.6%) compared to the non-invasive GTI and wound isolates (54.1% and 59.4%, respectively).
- •
For E. faecalis, agg was more frequent in isolates from the genital tract (57.1%) in comparison to the isolates from blood (22.2%) – p<0.05, while efaA was more common in isolates from UTI (94.3%) vs. from wound samples (72.9%) – p<0.01.
- •
For E. faecium, there were no significant differences in the distributions of genes encoding virulence factors, according to the infection site, but isolates from blood stream infections (n=8) were not considered due to their low number.
Prevalence of virulence genes in 510 clinical Enterococcus spp. (370 E. faecalis and 140 E. faecium) isolates according to the site of infection.
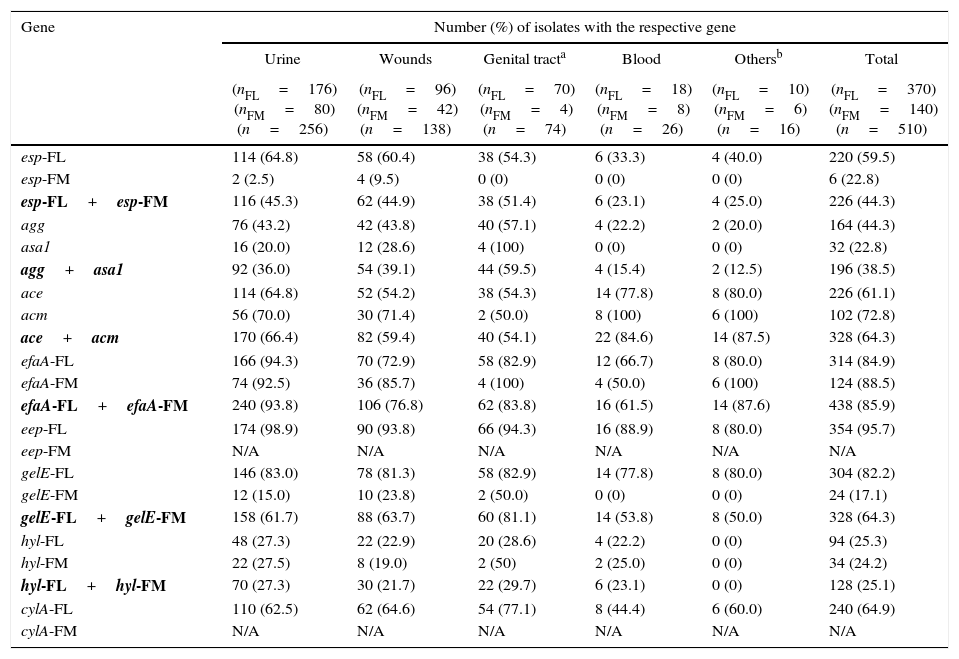
| Gene | Number (%) of isolates with the respective gene | |||||
|---|---|---|---|---|---|---|
| Urine | Wounds | Genital tracta | Blood | Othersb | Total | |
| (nFL=176) (nFM=80) (n=256) | (nFL=96) (nFM=42) (n=138) | (nFL=70) (nFM=4) (n=74) | (nFL=18) (nFM=8) (n=26) | (nFL=10) (nFM=6) (n=16) | (nFL=370) (nFM=140) (n=510) | |
| esp-FL | 114 (64.8) | 58 (60.4) | 38 (54.3) | 6 (33.3) | 4 (40.0) | 220 (59.5) |
| esp-FM | 2 (2.5) | 4 (9.5) | 0 (0) | 0 (0) | 0 (0) | 6 (22.8) |
| esp-FL+esp-FM | 116 (45.3) | 62 (44.9) | 38 (51.4) | 6 (23.1) | 4 (25.0) | 226 (44.3) |
| agg | 76 (43.2) | 42 (43.8) | 40 (57.1) | 4 (22.2) | 2 (20.0) | 164 (44.3) |
| asa1 | 16 (20.0) | 12 (28.6) | 4 (100) | 0 (0) | 0 (0) | 32 (22.8) |
| agg+asa1 | 92 (36.0) | 54 (39.1) | 44 (59.5) | 4 (15.4) | 2 (12.5) | 196 (38.5) |
| ace | 114 (64.8) | 52 (54.2) | 38 (54.3) | 14 (77.8) | 8 (80.0) | 226 (61.1) |
| acm | 56 (70.0) | 30 (71.4) | 2 (50.0) | 8 (100) | 6 (100) | 102 (72.8) |
| ace+acm | 170 (66.4) | 82 (59.4) | 40 (54.1) | 22 (84.6) | 14 (87.5) | 328 (64.3) |
| efaA-FL | 166 (94.3) | 70 (72.9) | 58 (82.9) | 12 (66.7) | 8 (80.0) | 314 (84.9) |
| efaA-FM | 74 (92.5) | 36 (85.7) | 4 (100) | 4 (50.0) | 6 (100) | 124 (88.5) |
| efaA-FL+efaA-FM | 240 (93.8) | 106 (76.8) | 62 (83.8) | 16 (61.5) | 14 (87.6) | 438 (85.9) |
| eep-FL | 174 (98.9) | 90 (93.8) | 66 (94.3) | 16 (88.9) | 8 (80.0) | 354 (95.7) |
| eep-FM | N/A | N/A | N/A | N/A | N/A | N/A |
| gelE-FL | 146 (83.0) | 78 (81.3) | 58 (82.9) | 14 (77.8) | 8 (80.0) | 304 (82.2) |
| gelE-FM | 12 (15.0) | 10 (23.8) | 2 (50.0) | 0 (0) | 0 (0) | 24 (17.1) |
| gelE-FL+gelE-FM | 158 (61.7) | 88 (63.7) | 60 (81.1) | 14 (53.8) | 8 (50.0) | 328 (64.3) |
| hyl-FL | 48 (27.3) | 22 (22.9) | 20 (28.6) | 4 (22.2) | 0 (0) | 94 (25.3) |
| hyl-FM | 22 (27.5) | 8 (19.0) | 2 (50) | 2 (25.0) | 0 (0) | 34 (24.2) |
| hyl-FL+hyl-FM | 70 (27.3) | 30 (21.7) | 22 (29.7) | 6 (23.1) | 0 (0) | 128 (25.1) |
| cylA-FL | 110 (62.5) | 62 (64.6) | 54 (77.1) | 8 (44.4) | 6 (60.0) | 240 (64.9) |
| cylA-FM | N/A | N/A | N/A | N/A | N/A | N/A |
FL, E. faecalis; FM, E. faecium; N/A, not applicable for E. faecium. esp, gene for enterococcal surface protein (adhesin) for both species; agg, E. faecalis aggregation substance involved in adherence to mammalian cells and conjugation; asa1, E. faecium aggregation substance involved in adherence to mammalian cells and conjugation; ace, E. faecalis Ace collagen-binding protein (adhesin); acm, E. faecium Acm collagen-binding protein (adhesin); efaA, cell wall adhesin expressed in patient serum (endocarditis-specific antigen A) for both species; eep (enhanced expression of pheromone), membrane protein of E. faecalis; gelE, extracellular metalloendopeptidase (gelatinase) for both species; hyl, hyaluronidase for both species; cylA, activation of cytolysin (hemolysin/class I. bacteriocin) of E. faecalis.
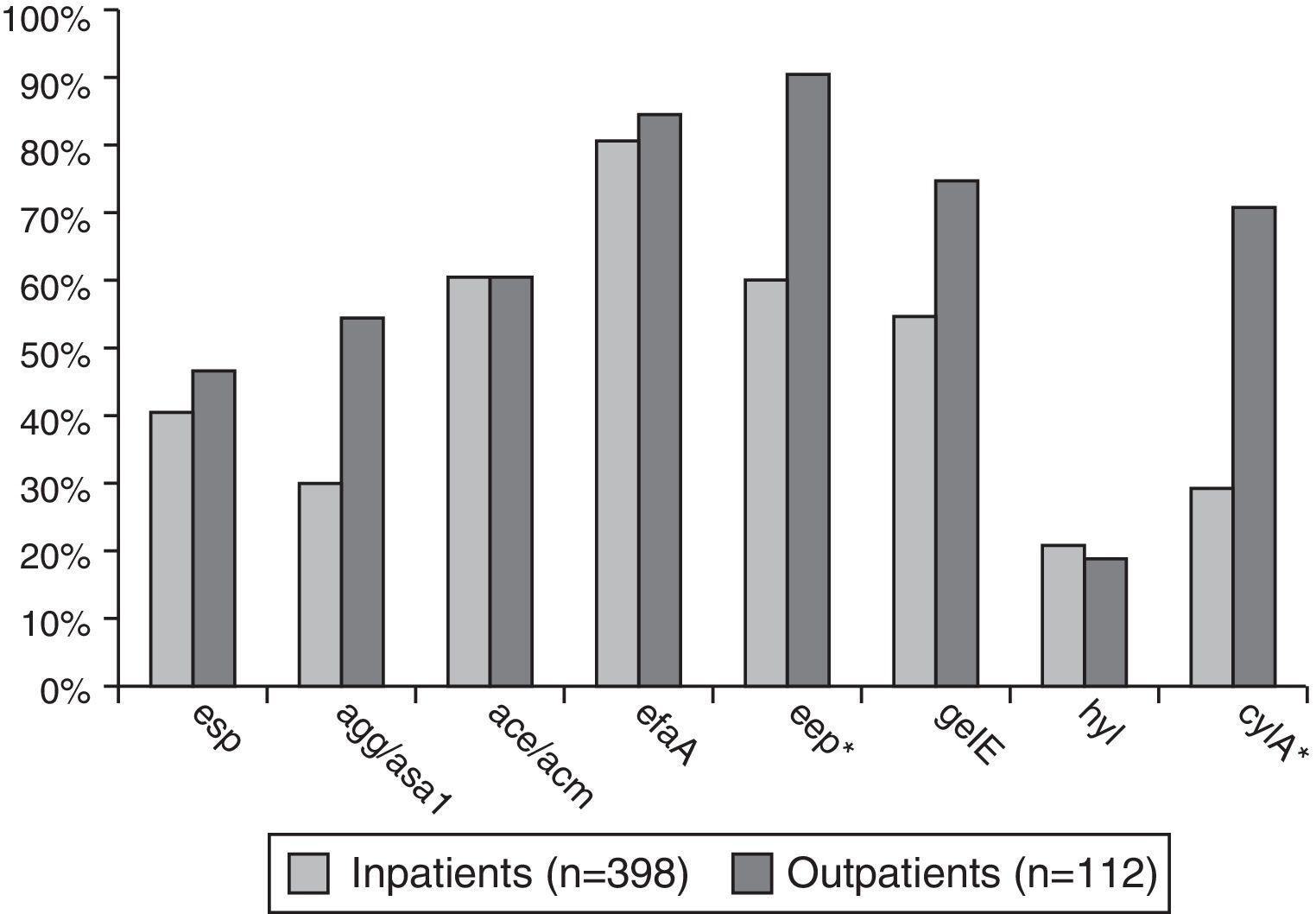
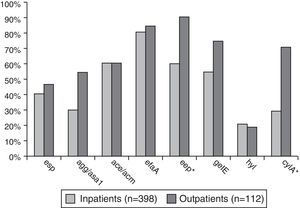
Enterococcus spp. isolates from outpatients had significantly higher prevalence (p<0.001–0.02) of the following virulence genes compared to the isolates from inpatients: agg/asa1 (58.9%/32.7%), eep (92.9%/62.8%), gelE (78.6%/60.3%), and cylA (73.2%/37.9%) – Fig. 2.
DiscussionIn this study, we found a significantly higher prevalence (p<0.001) of genes encoding some virulence factors involved in biofilm formation (the enterococcal surface protein (esp), aggregation substance (agg) and gelatinase (gelE)) in E. faecalis, compared to E. faecium. A previous study indicated higher prevalence of biofilm formation among E. faecalis strains compared to other enterococcal species.30E. faecalis has been recognized as the enterococcal species most commonly infecting indwelling medical devices.17,30 In a study conducted by Dupré et al. with clinically relevant enterococcal isolates from a hospital in Sardinia, Italy, in the period of 1998–2001 there was a complete absence of genes for gelatinase and aggregation substance in E. faecium, but contrary to our results, higher frequency of esp in E. faecium vs. E. faecalis was reported (71.9%/60%).23
The prevalence of efaA, eep, gelE, and cylA in E. faecalis isolates in our study was significantly higher than previously published data on clinical strains of E. faecalis from Brazil in 2004 – as it follows: 84.9%/58.9%; 95.7%/58.9%; 82.2%/45.3%; 64.9%/16.8% (p<0.001), while the gene frequency for agg and esp had similar rates in both studies (44.3%/36.8%, and 59.5%/57.9%).29 Detection of genes for adhesins (esp, agg, ace and efaA) in newer clinical E. faecalis isolates from Southern Brazil was more frequent than the rates found in our study (respectively, 68.4% in Brazil vs. 59.5% in our study, 57.9%/44.3%, 73.7%/61.1% and 100%/84.9%). In contrast, genes for gelE and cylA were more frequent in our isolates than among the Brazilian isolates (77.2%/82.2% and 54.4%/64.9%, respectively).31 A recent study of clinical isolates of E. faecalis in Italy showed a significantly lower frequency of cylA compared to our isolates (26.4%/64.9%, p<0.001), and close to our distribution of esp, agg, ace and efaA (52.7%/59.5%; 42.8%/44.3%; 67%/61.1%; 96.7%/84.9%).32
A multicenter study on the distribution of virulence determinants in fecal E. faecium isolates from patients in 13 hospitals from nine European countries showed total absence of asa1 and gelE, lower frequency of hyl in comparison to E. faecium isolates (17%/24.2%) of our study and significantly higher prevalence of esp (65% vs. 4.3%, p<0.001).26
The significantly higher prevalence of clinical E. faecalis isolates with multiple virulence determinants (over 4) compared to E. faecium, and the significant predominance of E. faecium isolates with one gene compared to E. faecalis, demonstrated in this study, was also recently found by Padmasini et al.33 According to another study of enteroccoci from clinical isolates and starter cultures, all strains of E. faecalis were characterized by a high number of virulence genes (between 6 and 11), while the E. faecium strains had no such gene determinants.25
In Italy, Creti et al. reported wider distribution of esp in non-invasive E. faecalis isolates (56.2%) compared to invasive isolates (38.4%).28 In our study esp was found in 33.3% of infections caused by E. faecalis, with bacteremia in the clinical course, while the frequency of esp in non-invasive infections varied from 54.3% (in GTIs) to 64.8% (in UTIs), which confirms the important role of Esp as a colonizing factor in UTIs.34 The frequency of esp among our UTI E. faecalis isolates (64.8%) was very similar to the rate reported by Creti et al. (66.7%). The findings for cylA in both studies were similar: 44.4% in invasive isolates from bacteremia in our data vs. 50% in the Italian study, as well as the results for cylA in non-invasive isolates – varying from 62.5% to 77.1% in our study and 62.5% in the Italian study.28
Recently Padilla et al. published their data on incidence of virulence genes amongst strains of E. faecalis isolated from UTIs and bacteremia in Chile, for the period of 2008–2009.35 The rates for the following genes from urine samples in our study were significantly higher than those reported in Chile: efaA (94.3%/62%, p<0.001), eep (98.9%/72%, p<0.001), gelE (83%/28%, p<0.001), cylA (62.5%/40%, p<0.01); while the presence of agg was significantly lower (43.2%/78%, p<0.001). Our isolates from bacteremia showed a significantly higher frequency of efaA compared to the Chilean isolates (66.7%/28%, p<0.001).
E. faecalis isolates from patients with UTIs and wound infections in the present study showed higher prevalence of numerous virulence genes, compared to similar isolates from Brazil, studied by Bittencourt de Marques and Suzart – esp (64.9% and 60.4% vs. 62% and 46.1%), agg (43.2% and 43.8%/38% and 38.5%), efaA (94.3% (p<0.001) and 72.9%/54% and 69.2%), eep (98.9% (p<0.001) and 93.8% (p<0.001)/58% and 53.8%), gelE (83% (p<0.001) and 81.3% (p<0.001)/50% and 34.6%) and cylA (62.5% (p<0.001) and 64.6% (p<0.001)/22% and 3.8%).29
An interesting finding from the present study was the higher dissemination rates of gene determinants for some virulence factors among Enterococcus spp. causing infections in outpatients, compared to those infecting inpatients. The review of published medical literature could not find a similar comparative study. Hospitalized patients may have a greater incidence of enterococcal infections not only because of virulence, but because the hospital itself is a risky hub. On the other hand, the intestinal tract is an important reservoir for opportunistic pathogens such as enterococci and allows them to access infected sites especially in outpatients through different virulence factors.21 A weakness of this comparative analysis results from the difficulties to obtain epidemiological data; often UTIs, occurring in inpatients, begin before admission to the hospital.
ConclusionsIn the present study clinical E. faecalis isolates were characterized by a significantly higher frequency of genes encoding some adhesins and gelatinase in comparison to E. faecium isolates. The higher virulence potential of E. faecalis vs. E. faecium isolates was proven (on the basis of a significantly higher prevalence of E. faecalis isolates with multiple virulence factor genes vs. E. faecium and significant predominance of E. faecium isolates without or with only one gene compared to the E. faecalis).
The genes for some adhesins (esp and agg/asa1) were more prevalent in non-invasive Enterococcus spp. isolates than in invasive isolates from bacteremia, while ace/acm showed a significantly higher frequency among invasive isolates.
For the first time, we proved that Enterococcus spp. isolates from outpatients with symptomatic infections had significantly higher frequency of the following virulence genes: agg/asa1, eep, gelE and cylA, in comparison to isolates causing infections in inpatients.
To the best of our knowledge, this is the first study in Bulgaria and in the Balkans region of the genes associated cellular and extracellular virulence factors among clinically relevant Enterococcus spp. isolates, in which a comparative analysis with respect to different features (species of microorganism, infection site, and origin) was made. Our results may serve as a basis for additional research of the pathogenesis of infections caused by Enterococcus spp.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by grants from the Medical University of Sofia, Bulgaria (Council of Medical Science, grant Nos. 16/2013 and 11/2014).