Human Bocavirus (HBoV) has been identified from feces and respiratory samples from cases of both acute gastroenteritis and respiratory illness as well as in asymptomatic individuals.
The aim of this study was to detect and characterize HBoV from fecal samples collected from hospitalized children aged less than five years old with no symptoms of respiratory tract infection (RTI) or acute gastroenteritis (AGE). The study involved 119 children and one fecal sample was collected from each participant between 2014 and 2015. HBoV was detected using Nested-PCR, and the viral type identified by genomic sequencing. HBoV-4 was identified from one sample obtained from a hospitalized child with soft tissue tumor of the submandibular region. This is the first report of HBoV-4 identification in Brazil, but we consider that this type may be circulating in the country similar to the other types and new investigations are necessary.
Human Bocavirus (HBoV), first described in 2005,1 belongs to the family Parvoviridae, genus Bocaparvovirus and includes four types (HBoV 1–4).2 The viral genome is composed of linear single-stranded DNA (ssDNA) of about 5kb3 with three open reading frames. The first and second regions encode the nonstructural proteins, NS1 and NP1, respectively, and the third encodes for VP1/VP2, which form the viral capsid.3 HBoV-1 has been linked to cases of respiratory tract infection (RTI), and types 2 and 3 are mainly detected in feces from children with acute gastroenteritis (AGE) or in asymptomatic cases for both syndromes.4,5 HBoV-4 has been less reported, but it has been identified in cases of RTI, AGE, and in healthy individuals.6–8 In Brazil, to our knowledge, types 1, 2, or 3 have been identified in children symptomatic for RTI or AGE.9–12 Moreover, to date HBoV-4 has not yet been identified in symptomatic or in healthy individuals in the country. This study aimed to detect and characterize HBoV in fecal samples from hospitalized children under five years old without RTI or AGE symptoms. This study presents the first identification of HBoV-4 in Brazil, detected in a child without symptoms of RTI or AGE who was hospitalized with tumor of the submandibular region.
Materials and methodsThe study was part of a cross-sectional study in the period 2014–2015 that involved children less than five years old, who were admitted to a child care referral hospital in Goiânia, Goiás – Hospital Materno Infantil. Respiratory (nasopharyngeal swab) and fecal samples were collected from each child. The group consisted of 192 children with RTI and/or AGE and 119 who were asymptomatic for both syndromes, and were hospitalized for other reasons like malnutrition, intestinal obstruction, a variety of surgeries, diabetes, heart diseases, kidney diseases, cancer, hematological diseases, liver diseases, and convulsive crisis. Clinical samples were collected only after children's parents or guardians completed and signed the Informed Consent Form (ICF) and provided their child's personal and demographic data. The study was approved by the Research Ethics Committee at the Clinical Hospital of Federal University of Goiás (protocol: 19920013.7.0000.5078). HBoV was identified by Nested-PCR, only from the fecal samples of the asymptomatic group, where viral DNA was extracted by commercial kit QIAamp® Viral RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions and primers specific for regions VP1 and VP2 were used as previously described.6 The first amplification reaction was performed using a commercial PCR mixture GoTaq (GoTaq Colorless Master Mix, Promega Corporation, WI, USA) and specific primers (20μM): AK-VP-F1 and AK-VP-R1. A second amplification (Nested-PCR) was performed with 1μL of the amplified product and the same reaction mixture and internal primers AK-VP-F2 and AK-VP-R2. Samples were considered positive when showed a fragment of 576bp. In all reactions, positive (previously sequenced archive HBoV sample) and negative (sterile water) controls were used in each run of samples.
Characterization of the positive samples was performed by genomic sequencing from product of the second amplification (576bp) that was purified with 65% isopropanol and 70% ethanol (Merck, Darmstadt, Germany), resuspended in formamide (Invitrogen, Foster, CA, USA) and denaturated (95°C for 5min). Reaction was performed according to a previously described method13 using the automatic sequencer ABI Prism3130 (Applied BiosystemsTM, USA), with at least two sequences in both directions generated for each sample. The procedure used 3μL purified DNA, 7μL of sequencing reagent Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA) and the primer pair of the second amplification.
In addition, a complete genome sequencing of positive samples was performed using the same method of the characterization step with the specific primers AK-VP-F1 and AK-VP-R16 as well as the primers previously described by Khamrin et al.14
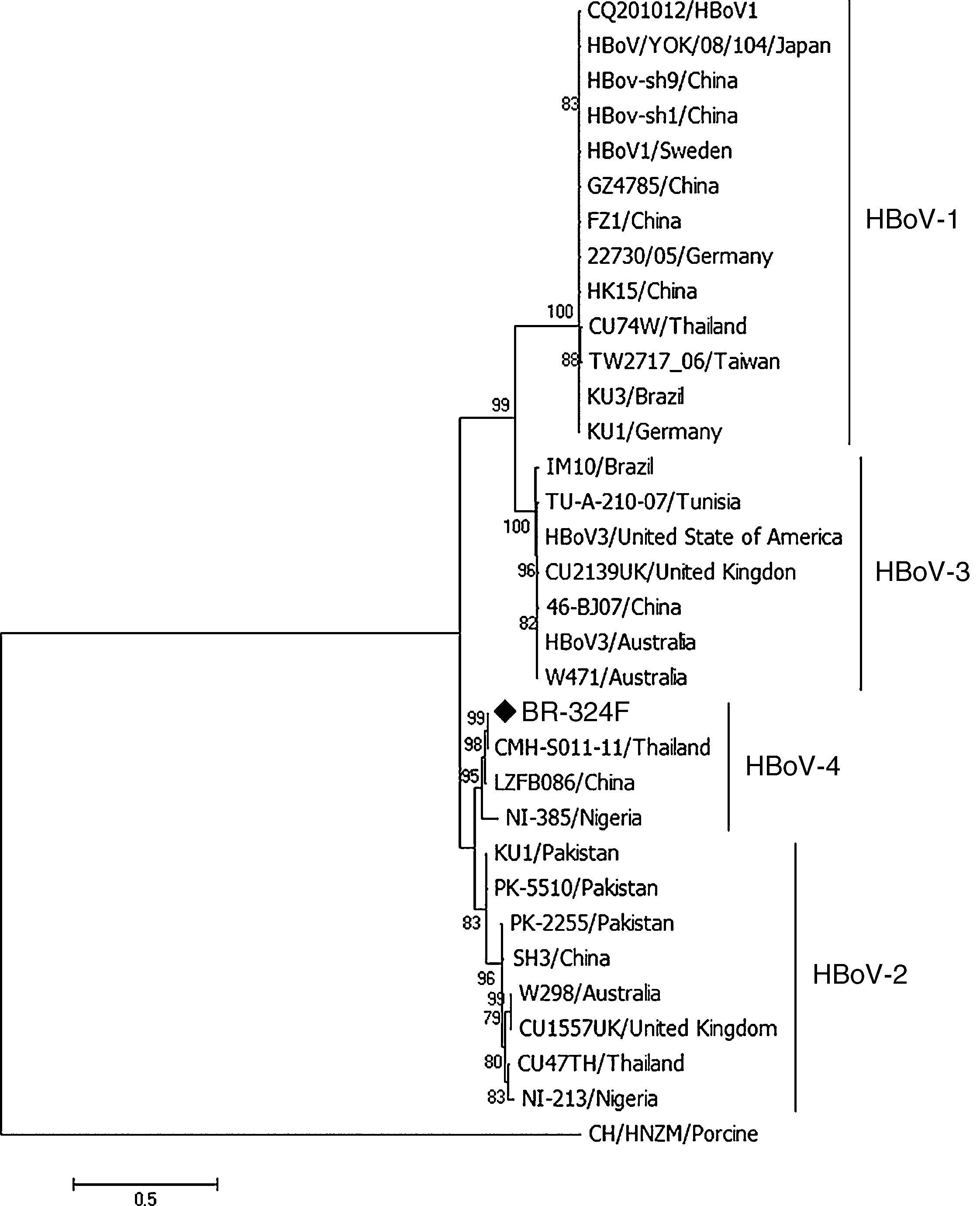
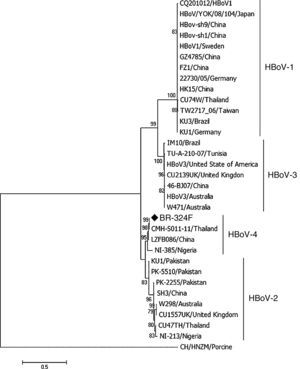
Phylogenetic analysis was performed using the program MEGA7 as described previously,15 considering 1000 bootstraps. The sequence of the complete genome of HBoV-4 has been deposited in GenBank under accession number KX826938.
ResultsFrom 119 fecal samples obtained from the asymptomatic group, one sample collected in September 2, 2015 was positive for HBoV-4. This sample was obtained from a 12-month old male child, who suffered from a soft tissue tumor of the submandibular region and had been hospitalized in the pediatric ward four days before the stool sample was collected. The HBoV-4 complete genome sequence showed high nucleotide identity to HBoV-4 samples from Thailand (99.6%), China (98.5%), and Nigeria (93.7%) (Fig. 1). Considering only coding regions, it was observed identity for NS1 (100.0%, 99.0% and 93.2%), NP1 (99.5%, 96.8% and 91.6%), and VP1/VP2 (99.6%, 99.1% and 95.6%) for samples from Thailand, China, and Nigeria, respectively. Analysis of NP1 coding sequence of HBoV-4 in relation to HBoV-1 and HBoV-3 samples from different parts of the world revealed up to 76.5% and 74.2% nucleotide identity to NP1 ORF of HBoV-1 and HBoV-3, respectively. For the HBoV-4 NS1 ORF, up to 73.8% identity was observed when compared to type 1, and 74.8% when compared to HBoV-3; for ORF VP1/VP2, up to 77.0% and 87.8% identities were observed for types 1 and 3, respectively (Fig. 2). When HBoV-4 sequence was compared to HBoV-2 sequences, the highest nucleotide identities were observed with respect to ORFs NP1 (97.3%) and NS1 (97.9%), whereas up to 87.1% similarity was observed for ORF VP1/VP2 (Fig. 2).
Phylogenetic analyses of the complete genome of HBoV-4 from feces of an individual asymptomatic for respiratory tract infection and acute gastroenteritis. The sample BR-324F is marked with a diamond. The scale bar corresponds to 0.5 nucleotide substitutions per site and the nodes have bootstrap values ≥70.
The observation of HBoV-4 detection in a child without RTI or AGE, who was hospitalized for a major morbidity, to our knowledge, is considered unique. HBoV-4 was first identified in 20106 in a multicenter study with fecal specimens. In that study, the virus was detected in six samples, four of which were from children who presented AGE and were suffering from symptoms of non-polio flaccid paralysis originating in Tunisia and Nigeria. The other two samples were from adults of the United States. One of these individuals had AGE and the other was considered healthy. Subsequent studies have reported the detection of HBoV- 4, such as those carried out by Khamrin et al., in Thailand7 and by Tymentsev et al. in Russia,16 where the agent was identified in fecal samples from one and two children who presented AGE, respectively. Another study by Koseki et al.8 also detected HBoV-4 from a nasopharyngeal swab obtained from a child with symptoms of RTI.8 The analysis of the HBoV-4 complete genome revealed high nucleotide identity with HBoV-4 samples from Southeast Asia and Africa, and the same was observed for the different ORFs of the virus genome. The analysis on the ORFs of HBoV-4 in relation to types 1, 2, and 3 showed great nucleotide divergence (>25%) for ORFs NP1 and NS1 of types 1 and 3, which is in line with the literature, with 87% of ORF VP1/VP2 identity between type 3 and HBoV-4.6,14 In contrast, high nucleotide identity (>80.0%) to type 2 was observed, considering all three ORFs.
In Brazil, to date, studies on HBoV have mainly been carried out in individuals with RTI17,18 or AGE,19,20 in which types 1, 2, or 3 were identified. Therefore, this is the first study to identify HBoV-4 in the country. The HBoV-4 sample was from a child hospitalized due soft tissue tumor of the submandibular region. Besides, studies from other countries have shown identification of HBoV-4 from cases of AGE or RTI as well as in healthy individuals.7,8 Our findings suggest that the agent could be associated with other clinical conditions. On the other hand, we consider that the infection of this child could have occurred due to the circulation of the virus in the community since that we have no evidence to indicate nosocomial infection. Thus, the virus is already circulating in Brazil, like other types, and new cases may be detected if suspected and investigated.
Conflicts of interestThe authors declare no conflicts of interest.
Funding for this research was provided by the Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG).