Burkholderia cenocepacia may cause serious infections in patients with cystic fibrosis, and this microorganism can be highly transmissible. Pulsed-field gel electrophoresis is widely used to study the dynamics of strain spread in cystic fibrosis patients. The aim of this work was to perform pulsed-field gel electrophoresis-based molecular typing of B. cenocepacia isolates to evaluate the epidemiology of this species at our hospital. A total of 28 isolates from 23 cystic fibrosis patients were analyzed. Initially, we compared isolates obtained from the same patient at different periods of time. We then compared the pulsed-field gel electrophoresis profiles of 15 IIIA isolates, and in a third analysis, evaluated the genetic profile of 8 IIIB isolates from different patients. The pulsed-field gel electrophoresis profiles of isolates from the same patient indicated that they are genetically indistinguishable. Analysis of isolates from different patients revealed the presence of multiple clonal groups. These results do not indicate cross-transmission of a unique clone of B. cenocepacia among cystic fibrosis patients, although this has been observed in some patients. Our findings highlight the importance of adequate patient follow-up at cystic fibrosis centers and adherence to management and segregation measures in cystic fibrosis patients colonized with B. cenocepacia.
Burkholderia cepacia complex (BCC) is a group of closely related Gram-negative bacteria that have emerged as opportunistic pathogens among individuals with cystic fibrosis (CF). BCC was described as 17 different species;1,2 however, studies have reported B. cenocepacia and B. multivorans as the predominant species in CF.3,4B. cenocepacia (formerly known as BCC genomovar III – phylogenetic subgroups IIIA, IIIB, IIIC and IIID) has been associated with rapid deterioration of lung function secondary to necrotizing pneumonia and sepsis, resulting in early death, among CF patients.5,6 This severe clinical condition, known as “cepacia syndrome,” was first recognized in the early 1980s, when a highly transmissible clone of B. cenocepacia IIIA (ET12) spread among CF patients in Canada, England and Scotland.7 Inter-patient transmission can occur during hospitalization or through social contact between CF patients outside the hospital setting.8 Therefore, some CF centers have begun to adopt policies for segregation of “B. cenocepacia-positive” and “negative” patients to prevent B. cenocepacia infection.4
Within this context, studies of the epidemiology of B. cenocepacia play an essential role in guiding infection control measures at CF centers. Consequently, the objective of this study was to perform molecular typing by DNA macrorestriction followed by pulsed-field gel electrophoresis (PFGE) of B. cenocepacia isolates in order to evaluate the epidemiology of this pathogen in CF patients at Hospital de Clínicas de Porto Alegre (HCPA), a tertiary-level university hospital with more than 800 beds in Porto Alegre, Brazil. The HCPA CF Reference Center provides care to nearly 250 CF patients. The hospital has a segregation program in place for BCC-colonized CF patients: colonized outpatients are seen on different days of the week than non-colonized CF patients, and colonized inpatients are admitted to isolation wards.
This study evaluated 28 B. cenocepacia isolates obtained from the sputum of 23 CF patients seen during the year 2008. During the same period, Leite and coworkers studied a total of 244 CF patients from HCPA and observed a prevalence rate of BCC of 10.6%. B. cenocepacia accounted for most BCC isolates (60%; 77.5% IIIA and 22.5% IIIB).9
Isolates were identified by phenotypic testing (characteristic growth on BCSA medium, polymyxin B resistance, pyrrolidonyl arylamidase [PYR] negativity) and with the miniAPI semi-automated ID/AST system, using the 32GN card (BioMérieux). B. cenocepacia identification was confirmed by nested PCR using primers for recA as described by Drevínek et al. B. cenocepacia was also used to identify phylogenetic subgroups IIIA and IIIB.10 The majority of our isolates (67.8%) were identified as genomovar IIIA (data not shown).
For PFGE analysis, DNA restriction was carried out with the SpeI enzyme and electrophoresis was performed in CHEF-DRII equipment (Bio-Rad Laboratories). Run conditions were as described by Kutty et al.11 Profiles were analyzed in the BioNumerics software environment (Applied Maths). The percentages of similarity of the test isolates were identified on a dendrogram derived from the UPGMA algorithm and based on the Dice coefficient. A similarity coefficient of 80% was used to define pulsed-field type clusters.12
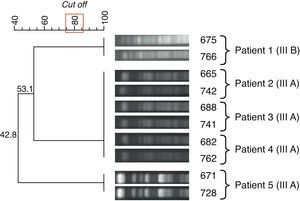
Three different analyses of isolates were performed on the basis of PFGE profiles. We first compared isolate profiles within the same patient (using a subsample of 5 patients and 10 isolates) at different points in time (4–6 months apart). These isolates showed the same DNA macrorestriction profile (100% genetic similarity – Fig. 1). On the basis of this finding, we decided to include only one isolate from each patient for analysis of isolate similarity among different patients.
The second analysis included 15 IIIA isolates from different patients. Nine different clonal groups (represented by letters A to I) were established (Fig. 2). Groups A, C, F, G, H and I included only one isolate; group B included 3 isolates that displayed 100% genetic similarity; group D included 4 isolates, 3 of which exhibited 100% similarity; and group E included 2 isolates also displaying 100% similarity. Notably, we found that the B. cenocepacia IIIA isolates from CF patients in our sample exhibited little genetic similarity with the ET12 strain.
The third analysis included 8 IIIB isolates from different patients and showed the presence of five distinct clonal groups (designated J to O) (Fig. 2). Groups L, N and O included only one isolate; group J included two isolates (710 and 768) that displayed 100% genetic similarity; and group M included 3 isolates (642, 656 and 706).
PFGE analysis revealed the presence of clonal groups formed by isolates with 100% genetic similarity and other groups with a similarity of >80%. We observed many different pulsotypes, most of which comprised only one isolate. These findings suggest the occurrence of multiple B. cenocepacia clonal groups among CF patients treated at HCPA, and, therefore, indicate absence of cross-transmission of a unique B. cenocepacia clone among CF patients, although this phenomenon has been observed in some patients (e.g. with clonal group D). Indeed, there were instances of different patients in the sample being colonized by the same strain.Several epidemiological studies have shown most cases of BCC infection transmitted between CF patients involved B. cenocepacia strains.13 The most dramatic clinical pictures were associated with the ET12 strain.14 Apparently, ET12 is an endemic strain isolated predominantly at CF centers in North America and Europe. However, our findings show that, on PGFE analysis, the DNA macrorestriction profile of ET12 strain isolates is quite distinct from that of B. cenocepacia IIIA isolates obtained from CF patients treated at HCPA.Our study also showed the persistence of particular genotype profiles in the same patients over time. In five patients, we observed the same B. cenocepacia clone in isolates collected 4–6 months apart. This suggests that, even during courses of antibiotic therapy, the original strain may not be eradicated from some patients.15 Genotyping studies have shown that chronic infection in CF patients is typically associated with a single BCC strain. Transient coinfection with two different BCC species or two strains of the same species may occur, but this phenomenon is rare and found mainly in early infection.16
In summary, we demonstrated the presence of multiple B. cenocepacia clonal groups among CF patients treated at Hospital de Clínicas de Porto Alegre. Our findings highlight the importance of adequate patient follow-up at CF centers and adherence to management and segregation measures in CF patients colonized with B. cenocepacia.
Conflict of interestThe authors have no conflicts of interest to declare.
The authors would like to honor the memory of Ms. Maria Izolete Vieira (Unidade de Microbiologia e Biologia Molecular – Hospital de Clínicas de Porto Alegre) and acknowledge her technical support. We also thank Ms. Odelta dos Santos (Universidade Federal de Ciências da Saúde de Porto Alegre – UFCSPA) for her assistance with BioNumerics software analysis. This work was supported by Fundo de Incentivo à Pesquisa e Ensino, Hospital de Clinicas de Porto Alegre (FIPE/HCPA), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and the Brazilian National Council for Scientific and Technological Development (CNPq).