Several studies have been conducted in recent years to elucidate the structure, function and significance of AcrB, MarA, SoxS and RamA in Salmonella enterica. In this study, the relative quantification of acrB, soxS, marA and ramA genes expression was evaluated in 14 strains of S. enterica, with or without accompanying mutations in the quinolone resistance-determining regions of the gyrA gene, that were exposed to ciprofloxacin during the exponential growth phase. The presence of ciprofloxacin during the log phase of bacterial growth activated the genes marA, soxS, ramA and acrB in all S. enterica strains analyzed in this study. The highest expression levels for acrB were observed in strains with gyrA mutation, and marA showed the highest expression in the strains without mutation. Considering only the strains with ciprofloxacin minimum inhibitory concentration values<0.125μg/mL (sensitive to ciprofloxacin), the most expressed gene in the strains both with and without mutations was acrB. In the strains with ciprofloxacin minimum inhibitory concentration values≥0.125μg/mL (low susceptibility), with and without mutations in gyrA, the most expressed gene was marA. In this study, we observed that strains resistant to nalidixic acid may express genes associated with the efflux pump and the expression of the AcrAB-TolC pump genes seems to occur independently of mutations in gyrA.
The resistance of Salmonella enterica to quinolones is usually mediated by mutations in topoisomerase genes. However, the presence of plasmids and active efflux pumps can be determinant factors in decreased antibiotic susceptibility and increased resistance.1
Efflux pumps act in response to bacterial stress and the expression of pumps reduces the accumulation of organic compounds or antimicrobial agents within the cell and can also be caused by chromosomal mutations, resulting in multiple antibiotic resistance (MAR).2 According to Baucheron et al.,3 efflux pumps play an important role in resistance, especially when fluoroquinolones are involved.
The AcrB efflux pump belongs to the resistance-nodulation-cell division (RND) superfamily and acts in association with two types of proteins: the outer membrane channel (TolC) and a periplasmic ‘adaptor’ protein (AcrA).4 Regulators belonging to the AraC/Xyls family of transcriptional activators,5 i.e. marA,6soxRS,7rob,8 and ramA9 have been found to activate acrAB transcription by binding to the marbox sequence characterized in its promoter.10
The MarA and SoxS proteins can activate acrAB expression. The marRAB operon is responsible for producing the MarA transcriptional activator protein,6 which can be synthesized in response to the presence of antibiotics and can result in the multi-drug resistance phenotype (MDR). In turn, the soxRS operon is activated in response to oxidative stress, with the SoxS protein subsequently acting as transcriptional activator.11 The overexpression of soxRS also contributes to increased antibiotic resistance in Gram-negative bacteria.
The product of the ramR gene regulates the gene ramA,12 which produces the protein RamA. RamA is homologous to MarA and SoxS.13 Bacteria overexpressing ramA may exhibit the MDR phenotype by inducing expression of acrAB and tolC.9
S. enterica strains with the tolC gene experimentally deleted show an increased susceptibility to antimicrobial agents.14 Similarly, strains of S. enterica showed higher susceptibility to ciprofloxacin when ramA was inactivated.15
Although several studies have been conducted in recent years to elucidate the structure, function and significance of AcrB, MarA, SoxS and RamA,16–18 their regulation in S. enterica is not fully understood. In this study, the expression of acrB, soxS, marA and ramA was evaluated in fourteen strains of S. enterica, with or without accompanying mutations in the quinolone resistance-determining regions (QRDR) of the gyrA gene, that were exposed to ciprofloxacin during the exponential growth phase.
Materials and methodsStrainsWe analyzed 14 strains of S. enterica that are resistant to nalidixic acid (NAL). These strains were divided into two groups: seven strains without mutation in the QRDR region of the genes gyrA, gyrB, parC and parE and seven strains with mutations in the QRDR of gyrA, but with no mutations in the QRDR of other genes. Among the strains with mutations in the gyrA QRDR, one was positive for the qnrB19 plasmid gene. The ciprofloxacin minimum inhibitory concentration (CipMIC) of each strain was determined using E-test gradient strips (AB Biodisk, Solna, Sweden). Once the CipMIC had been determined, each strain was cultivated in duplicate for 2h in Mueller-Hinton broth (MHB) and ciprofloxacin was added to one of the two cultures at a concentration corresponding to half of the correspondently CipMIC of each strain.19 After 30min of exposure to the antibiotic at 37°C, the culture was centrifuged for subsequent RNA extraction.
RNA isolationRNA isolation was performed according to Chomczynski and Sacchi20 by the guanidinium-thiocyanate–phenol–chloroform method, with some modifications. After centrifugation at 14,000×g for 10min, the supernatant was discarded, 200μL of lysis solution was added and the samples were incubated for 1h at 37°C. After this time, 50μL of 1% SDS was added. After vortex agitation, 25μL of 2M NaCl and 200μL phenol:chloroform were mixed into the material; which was placed on ice for 10min. The material was then centrifuged at 14,000×g for 20min at 4°C, and the supernatant was recovered. An equal volume of isopropanol was added to the supernatant; mixture was incubated at −70°C for 2h and then centrifuged at 14,000×g for 11min at 4°C. After the supernatant was discarded, the pellet was dried under air flux and then vortexed in 750μL of 75% ethanol. A new centrifugation was conducted at 14,000×g for 11min at 4°C; the supernatant was discarded; the pellet was dried for 1h and resuspended in 30μL of nuclease-free water (Invitrogen™). The RNA samples were treated with RQ1 DNase (Ambion AM 1906), following the manufacturer's instructions. RNA purity and concentration was measured using the Nanodrop ND-1000.
Reverse transcriptase (RT)RT reactions were conducted as described by Chico et al.,21 using 1μg of total RNA, 90ng of random hexamers (QIAGEN®) and 0.5mM dNTPs (Invitrogen™). Prior to cDNA synthesis, denaturation was performed for 5min at 65°C. After this step, 200U of MMLV reverse transcriptase (Invitrogen™), 10mM of DTT (Invitrogen™), and 20 units of RNase inhibitor (RNase OUT, Invitrogen) were added to each reaction to a final volume of 20μL. The reaction conditions were 10min at 25°C, 15min at 42°C and 5min at 99°C.
Relative quantification of gene expressionAfter obtaining the cDNA, quantitative real-time reverse transcriptase PCR (qRT-PCR) was performed using the 7500 Real-Time PCR System, SDS software version 1.4 (Applied Biosystems). The reactions were performed in a final volume of 20μL, containing 300nM of each primer, 100nM of each probe (Table 1), 2μL of cDNA and 1× TaqMan® universal master mix (Applied Biosystems). Standard curves were constructed to calculate the gene-specific PCR efficiency from 10-fold series dilution of the mixed cDNA template for each primer pair. The efficiency of qRT-PCR reactions was calculated using the formula E=[10(−1/S)]−1, where E represents the calculated efficiency, and S is the slope of the standard curve.22 To check all primers specificity, real-time PCR was performed on cDNA and products were analyzed by electrophoresis on 2% agarose gel and ethidium bromide staining. Negative qRT-PCR control with no templates was performed for each primer pair.22
TaqMan® primers and probes. Sequences of primers and probes used for qRT-PCR (quantitative real-time reverse transcriptase PCR) reactions, designed with the help of the Primer Express™ 2.0 software (Applied Biosystems).
| Primers and probesa | Primers sequences 5′→3′ |
|---|---|
| 16s-F | CGTGTTGTGAAATGTTGGGTTAA |
| 16s-R | CCGCTGGCAACAAAGGATAA |
| probe 16s | TCCCGCAACGAGCGCAACC |
| acrB-F | TGAAGACCAGGGCGTATTCCT |
| acrB-R | TTTTTGCGTGCGCTCTTG |
| probe acrB | ACAATGGTCCAGCTCCCCGCG |
| soxS-F | CGGAATACACGCGAGAAGGT |
| soxS-R | GAGCGCCCGATTTTTGATATC |
| probe soxS | TGCTGCGATACATAGCCCAGGTCCA |
| marA-F | GACCCGGACGTTCAAAAACTAT |
| marA-R | TCGCCATGCATATTGGTGAT |
| probe marA | TGATGTGCCGCCACACAAATACCG |
| ramA-F | CCAGAAGGTGTATGATATTTGTCTCAAG |
| ramA-R | GGTTGAACGTGCGGGTAAA |
| probe ramA | TTGATTCGCAGCAAACCTTTACGCG |
The results were analyzed using SPSS 18.0 software. The geometric means (GM) of the CipMICs were calculated using the following y1y2y2⋯ynn formula: where y represents the individual CipMIC of each strain, and n represents the total number of CipMIC values.23
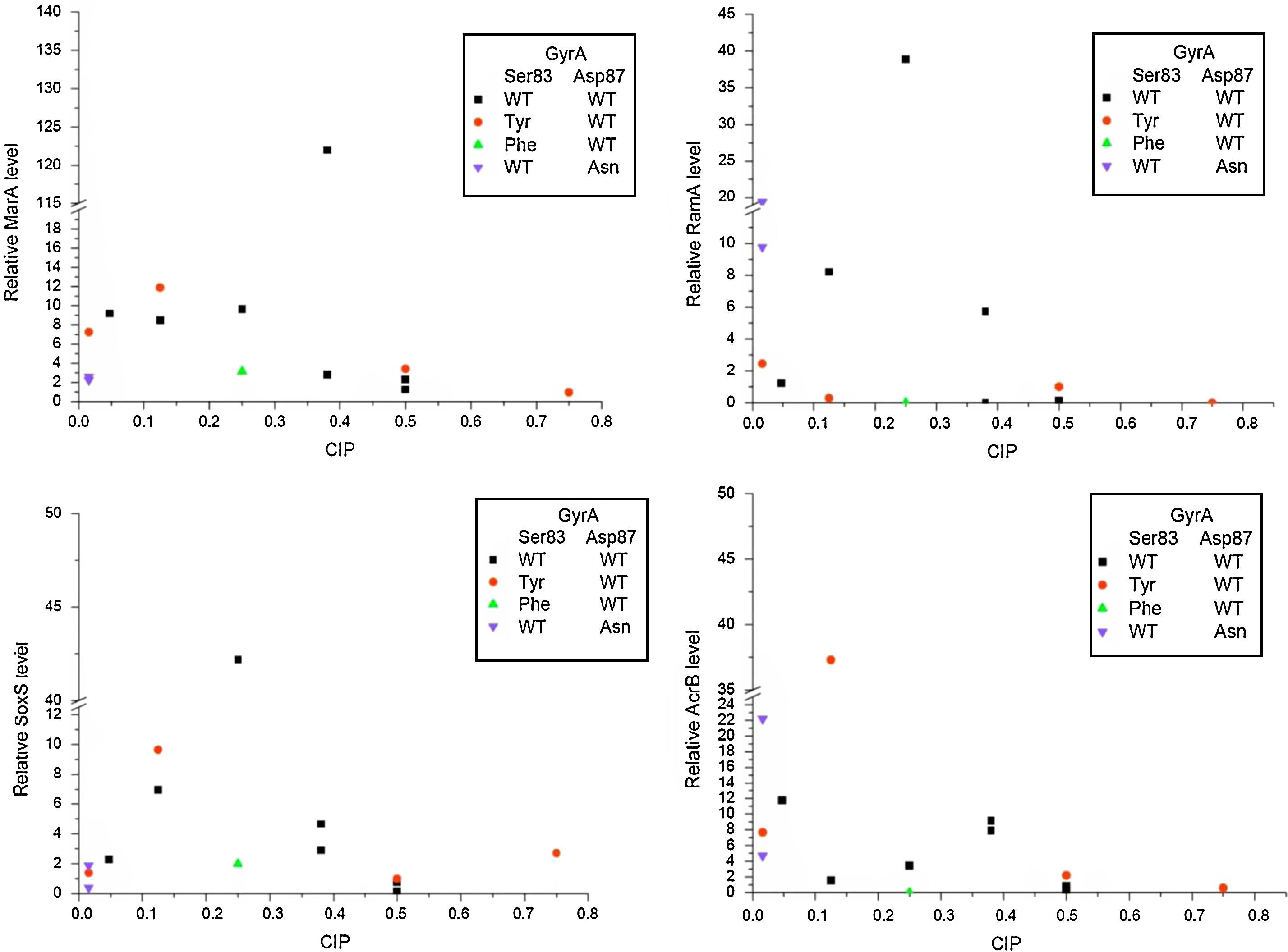
ResultsThe presence of ciprofloxacin during the log phase of bacterial growth activated at least one of the four genes tested in all S. enterica strains analyzed in this study. The expression levels of the genes marA, soxS and ramA were higher in the strains without a mutation in gyrA, gyrB, parA or parC than for strains with mutations. However, the highest expression levels for acrB were observed in strains with gyrA mutation (Fig. 1).
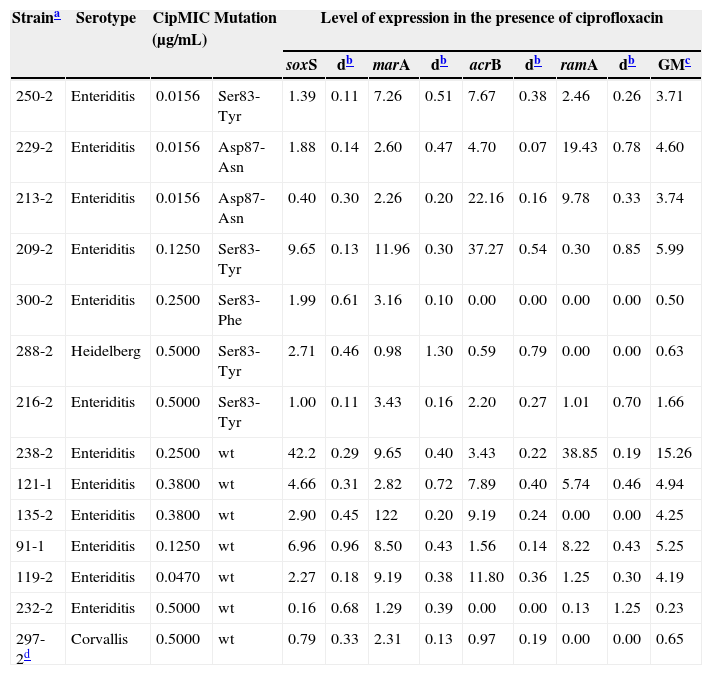
The GM value for the four pump genes evaluated in Salmonella strains without mutations was 2.70, and for strains with a mutation in gyrA it was 2.13. Among the strains mutated in gyrA, strain 209-2 showed higher expression of the genes analyzed, with a 5.99GM. In contrast, the strain without mutation in gyrA with the highest levels of gene expression was 238-2, with a 15.38GM (Table 2).
Distribution of the mutations observed in QRDR of the gyrA gene in Salmonella strains of different serovars with the corresponding minimal inhibitory concentration for ciprofloxacin (CipMIC) and geometric mean (GM) data.
| Straina | Serotype | CipMIC (μg/mL) | Mutation | Level of expression in the presence of ciprofloxacin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| soxS | db | marA | db | acrB | db | ramA | db | GMc | ||||
| 250-2 | Enteriditis | 0.0156 | Ser83-Tyr | 1.39 | 0.11 | 7.26 | 0.51 | 7.67 | 0.38 | 2.46 | 0.26 | 3.71 |
| 229-2 | Enteriditis | 0.0156 | Asp87-Asn | 1.88 | 0.14 | 2.60 | 0.47 | 4.70 | 0.07 | 19.43 | 0.78 | 4.60 |
| 213-2 | Enteriditis | 0.0156 | Asp87-Asn | 0.40 | 0.30 | 2.26 | 0.20 | 22.16 | 0.16 | 9.78 | 0.33 | 3.74 |
| 209-2 | Enteriditis | 0.1250 | Ser83-Tyr | 9.65 | 0.13 | 11.96 | 0.30 | 37.27 | 0.54 | 0.30 | 0.85 | 5.99 |
| 300-2 | Enteriditis | 0.2500 | Ser83-Phe | 1.99 | 0.61 | 3.16 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.50 |
| 288-2 | Heidelberg | 0.5000 | Ser83-Tyr | 2.71 | 0.46 | 0.98 | 1.30 | 0.59 | 0.79 | 0.00 | 0.00 | 0.63 |
| 216-2 | Enteriditis | 0.5000 | Ser83-Tyr | 1.00 | 0.11 | 3.43 | 0.16 | 2.20 | 0.27 | 1.01 | 0.70 | 1.66 |
| 238-2 | Enteriditis | 0.2500 | wt | 42.2 | 0.29 | 9.65 | 0.40 | 3.43 | 0.22 | 38.85 | 0.19 | 15.26 |
| 121-1 | Enteriditis | 0.3800 | wt | 4.66 | 0.31 | 2.82 | 0.72 | 7.89 | 0.40 | 5.74 | 0.46 | 4.94 |
| 135-2 | Enteriditis | 0.3800 | wt | 2.90 | 0.45 | 122 | 0.20 | 9.19 | 0.24 | 0.00 | 0.00 | 4.25 |
| 91-1 | Enteriditis | 0.1250 | wt | 6.96 | 0.96 | 8.50 | 0.43 | 1.56 | 0.14 | 8.22 | 0.43 | 5.25 |
| 119-2 | Enteriditis | 0.0470 | wt | 2.27 | 0.18 | 9.19 | 0.38 | 11.80 | 0.36 | 1.25 | 0.30 | 4.19 |
| 232-2 | Enteriditis | 0.5000 | wt | 0.16 | 0.68 | 1.29 | 0.39 | 0.00 | 0.00 | 0.13 | 1.25 | 0.23 |
| 297-2d | Corvallis | 0.5000 | wt | 0.79 | 0.33 | 2.31 | 0.13 | 0.97 | 0.19 | 0.00 | 0.00 | 0.65 |
wt, strain without mutations in gyrA, gyrB, parC and parE.
After assessing the level of expression for each gene in the strains without mutation, marA showed the highest expression (GM=6.93), followed by soxS (GM=2.72), acrB (GM=2.38) and ramA (GM=1.16). In the strains with mutations in gyrA, the gene marA also showed higher levels of gene expression (GM=3.41) but was followed by acrB (GM=3.25), soxS (GM=1.77) and ramA (GM=1.05).
Considering only the strains with CipMIC values<0.125μg/mL (sensitive to ciprofloxacin), the most expressed gene in the strains both with and without mutations was acrB. In the three mutated strains, the GM for this gene was 9.28, and in the strain without mutation the level of expression of acrB was 11.8 (Table 2).
In the strains with CipMIC values≥0.125μg/mL (low susceptibility), with and without mutations in gyrA, the most expressed gene was marA. For the four mutated strains, marA expression ranged from 0.98 to 11.96, and the GM was 3.36. In contrast, for the six strains without mutation, the marA expression level ranged from 1.29 to 122, with a GM of 6.62.
Among the four strains with gyrA mutations and reduced susceptibility to ciprofloxacin, strain 288-2, serovar Heidelberg, and strain 300-2, serovar Enteritidis, scarcely expressed the pump genes tested. On the other hand, S. Enteritidis strain 209-2, despite a GM of 5.99, presented a CipMIC value four times lower than S. Enteritidis strain 216-2, which showed a GM of 1.66 (Table 2).
Four of the six strains without mutations and reduced susceptibility to ciprofloxacin showed significant expression of the efflux pump genes evaluated, with GM values between 4.25 and 15.26. In contrast, strain 232-2, with no mutation in gyrA and the lowest level of gene expression linked to the pump (GM=0.23), presented a CipMIC value of 0.5μg/mL. Strain 297-2, serovar Corvallis, positive for the qnrB19 plasmid gene, also presented no expression of any of the four genes studied that are linked to the pump complex (GM=0.65).
DiscussionThere are reports that AcrAB pump expression in Escherichia coli can increase the MIC for ciprofloxacin and norfloxacin up to five times.24 In this study, all the strains evaluated showed expression of at least one gene related to efflux pumps in the presence of ciprofloxacin. However, the expression of these genes and their relationship with reduced susceptibility to ciprofloxacin was highly divergent, independent of mutations in gyrA. The S. Enteritidis strains 250-2, 229-2 and 213-2, with mutations in gyrA and expression of at least 3 genes linked to the pump, presented CipMIC values of less than 0.125μg/mL.
However, other S. Enteritidis strains (209-2, 300-2 and 216-2), with gyrA mutations and both with and without expression of the genes studied, showed reduced susceptibility to ciprofloxacin.
Baucheron et al.3 have reported a reduction of up to 64-fold in CipMIC values after inhibition of the AcrAB operon in strains of S. Typhimurium, with or without mutations in gyrA. Morgan-Linnell et al.25 also failed to correlate AcrAB expression and the presence of mutations in topoisomerase genes with the MIC for fluoroquinolones. The increase in the levels of AcrAB expression did not always increase the MICs, suggesting that isolated AcrAB expression does not generate high MIC values. Furthermore, the expression of the efflux pump was reported as being responsible only for baseline resistance.26
In this study, strains of S. Enteritidis without QRDR mutations in the genes gyrA, gyrB, parC or parE showed reduced susceptibility to ciprofloxacin. The expression of genes associated with the efflux pump seems to have influenced this reduction. Strains 121-1, 135-2, 91-1 and 238-2 of S. Enteritidis showed a GM of 4.94, 4.25, 5.25 and 15.26, respectively. In this group, it is important to note that strain 135-2 presented marA expression of 122, and strain 238-2 of S. Enteritidis increased expression of both the soxS and ramA genes as well as marA (Table 2).
The expression of marA is induced by a number of substances, including quinolones.27 The marA gene is able to mediate drug resistance by decreasing expression of the OmpF porin and causing overexpression of the AcrB efflux pump.6,16 Studies with E. coli reported MIC values for ofloxacin up to eightfold higher in strains with overexpression of the marA gene, in comparison to strains that only had mutations in gyrA.12,28,29 Kern et al.30 observed that the rate of successful treatment of E. coli with fluoroquinolones was significantly lower in strains that expressed marA. In clinical isolates, increased levels of AcrAB pump gene expression may arise due to changes in marA or to bactericidal protection generated by the regulators of this gene. The fact that the marA gene is associated with the marRAB operon activator suggests its use (marA) as a target for studies on strategies against microbial resistance.25
The soxS gene was the most expressed, after marA, in the non-mutated strains analyzed in this study. This gene is the transcription activator that positively regulates the expression of more than 20 genes related to the increased synthesis of proteins associated with the AcrAB-TolC efflux pump.31–33 The SoxRS regulatory system protects cells against oxidative stress generated by various antimicrobials.32 The S. Enteritidis strain 238-2, without mutations in gyrA, showed expression of the soxS gene and reduced susceptibility to ciprofloxacin. Kehrenberg et al.34 observed an eightfold increase in the CipMIC values of S. Virchow strains when soxS was expressed but no relationship with the presence of gyrA mutations.
The expression of efflux pump genes was practically zero for the Heidelberg serovar strain (GM=0.63), which showed the highest CipMIC among all the strains analyzed in this study. It suggests that this reduction in susceptibility is due to the intrinsic resistance commonly observed in this serovar.35
The Corvallis serovar presented a CipMIC of 0.5μg/mL, but no mutation in the QRDR region of the topoisomerase genes that correlates with the low expression levels for the studied genes related to efflux pumps. However, the reduced susceptibility to ciprofloxacin observed may relate to the presence of the qnrB19 plasmid gene. According to Li,36 the qnr gene facilitates the selection of chromosomal mutations related to quinolone resistance in S. enterica by significantly increasing the levels at which mutants can be selected.
The 232-2 S. Enteritidis strain presented a CipMIC of 0.5μg/mL and did not express any of the studied genes linked to the efflux pump. It is possible that alternative mechanisms of gene regulation, such as acrR, that are independent of mar-sox-rob were responsible for controlling the expression of AcrB in S. enterica.37 Mutations in genes controlling the transcription or expression of other genes, such as acrD, acrA or tolC15 may also be involved, in addition to the AcrEF efflux previously described in S. enterica.38,39
Another possibility for the reduced susceptibility to ciprofloxacin observed in strain 232-2 could be mutations in gyrA outside the QRDR region. Capoor et al.40 have suggested an increase to the regional boundaries that include the QRDR of the gyrA gene because mutations associated with resistance to quinolones were observed beyond the current domain.
In this study, we observed that strains resistant to NAL may express genes associated with the efflux pump as a form of primary resistance to this drug. This mechanism may perform a pre-selection for less sensitive strains, with the subsequent emergence of mutated and resistant strains. One possible mechanism to increase the life of quinolones and fluoroquinolones would be the development of inhibitors of the AcrAB-TolC system, which could minimize the selection of strains resistant to NAL. The expression of the AcrAB-TolC pump genes seems to occur independently of mutations in gyrA, because strains without mutations in the QRDR regions of the topoisomerase genes evaluated in this study presented higher expression of the genes associated with the efflux pumps. However, the results indicate the need for additional studies to better understand the complex interplay between the expression and regulation of efflux pump genes and other mechanisms of resistance to quinolones in S. enterica.
Conflict of interestAll authors declare to have no conflict of interest.
We would like to acknowledge CNPq and Capes for financial support in the form of PhD scholarships awarded to RF and MM.