Streptococcus agalactiae is a common agent of clinical and subclinical bovine mastitis and an important cause of human infections, mainly among pregnant women, neonates and nonpregnant adults with underlying diseases. The present study describes the genetic and phenotypic diversity among 392 S. agalactiae human and bovine strains isolated between 1980 and 2006 in Brazil. The most prevalent serotypes were Ia, II, III and V and all the strains were susceptible to penicillin, vancomycin and levofloxacin. Resistance to clindamycin, chloramphenicol, erythromycin, rifampicin and tetracycline was observed. Among the erythromycin resistant strains, mefA/E, ermA and, mainly, ermB gene were detected, and a shift of prevalence from the macrolide resistance phenotype to the macrolide-lincosamide-streptogramin B resistance phenotype over the years was observed. The 23 macrolide-resistant strains showed 19 different pulsed-field gel electrophoresis profiles. Regarding macrolide resistance, a major concern in S. agalactiae epidemiology, the present study describes an increase in erythromycin resistance from the 80s to the 90s followed by a decrease in the 2000–2006 period. Also, the genetic heterogeneity described points out that erythromycin resistance in Brazil is rather due to horizontal gene transmission than to spreading of specific macrolide-resistant clones.
Streptococcus agalactiae (Group B Streptococcus; GBS) is a common agent of clinical and subclinical bovine mastitis, accounting for great economic losses in dairy industry worldwide.1,2 Additionally, it is an important source of severe diseases in newborns and pregnant women, and an emerging cause of invasive infections in nonpregnant adults, mostly in those presenting underlying conditions.3,4
Asymptomatic colonization can evolve to invasive diseases in susceptible human hosts; in animal hosts, non-treated subclinical mastitis usually turns into chronic cases leading to permanent damages.2,4 For this reason, high-quality prevention and early treatment are crucial goals, which are generally achieved through the usage of antimicrobials. In cattle, commercial solutions (mostly including macrolides, penicillins and tetracyclines) are usually employed as one of the mastitis control strategies.2,5 In humans, penicillin remains the drug of choice for chemoprophylaxis according to Centers of Disease Control (CDC).6
Macrolides and lincosamides represent the alternative in cases of therapeutic failure and of patients allergic to beta-lactams, although human and bovine GBS strains resistant to those drugs have been described in the last decades, varying from 2.0% to 54.0%.7,8 Likewise, rates of tetracycline-resistant GBS isolates from human and bovine hosts have been increasing.8–10
Ten S. agalactiae capsular types were described so far (Ia, Ib, II–IX), and serotyping is still the first epidemiological approach to characterize GBS strains.11 In addition, pulsed-field gel electrophoresis (PFGE) has been used to evaluate the genetic diversity and clonal relatedness within this species.8,12,13 In Brazil, few data about the genetic diversity, serotyping and antimicrobial susceptibility of S. agalactiae are available. Therefore, here we provide longitudinal data about distribution of capsular types, profiles of antimicrobial susceptibility, including major mechanisms of erythromycin resistance, and genetic diversity of erythromycin resistant strains among S. agalactiae isolated from humans and cattle in Brazil.
Materials and methodsBacterial strainsA total of 392 GBS strains, including 363 from humans and 29 from bovine origin, were analyzed. One hundred forty three strains were isolated between 1980 and 1989, 87 between 1990 and 1999 and 162 between 2000 and 2006. Most of the isolates were recovered during different studies performed by our group, including 289 isolates previously serotyped3,14–16 and were stored at −20°C in our collection of cultures.
The human originated strains were isolated between 1980 and 2006 in the States of Rio de Janeiro, Santa Catarina, and São Paulo, from patients and healthy carriers, including children, pregnant women and nonpregnant adults, and were mainly from throat (n=69), urine (n=58), vagina (n=51), anus (n=22), cervix (n=18), outer ear (n=9) and liquor (n=7). Others less common clinical origins included blood (n=4), surgical wounds (n=4), peritoneum (n=1), skin (n=3), placenta (n=4), lungs (n=2), sperm (n=4) and urethra (n=1). The bovine originated strains were isolated from milk of clinical or subclinical mastitis cases at 5 different herds in the States of Rio de Janeiro, Santa Catarina, and São Paulo, between 1987 and 1989 and between 2003 and 2006.
Determination of capsular serotypesOne hundred and three strains isolated between 2001 and 2002 in Rio de Janeiro from human hosts were serotyped by immunoprecipitation using specific antisera for types Ia, Ib, and II–VIII, which were prepared in house using recognized reference strains.3
Antimicrobial susceptibility testingAntimicrobial susceptibility testing of 392 strains was performed by Kirby–Bauer method using clindamycin, chloramphenicol, erythromycin, levofloxacin, penicillin, rifampicin, tetracycline and vancomycin disks (Cecon, São Paulo, Brazil). Streptococcus pneumoniae ATCC 49619 was used as quality control and CLSI breakpoints as the interpretative criteria.17
Determination of erythromycin resistance phenotypes and genotypesThe erythromycin-clindamycin double-disk test was carried out for the determination of the resistance phenotypes among all the erythromycin resistant strains, as previously described.18 The presence of ermA, ermB, mefA/E and lnuB genes was also investigated among these strains using polymerase chain reaction (PCR). DNA extraction was performed according to Sambrook et al.19 The reactions were performed in a GeneAmp PCR System 2400 (Applied Biosystems) using primers and cycles previously described.20–22 PCR-amplified products were run on 1% agarose gels and stained with ethidium bromide. The 100-bp DNA ladder kit (Invitrogen) was used as the DNA size marker.
Pulsed-field gel electrophoresis (PFGE) analysisPFGE profiles from all erythromycin resistant strains were obtained as previously described.23 The genomic DNA was digested with the SmaI restriction enzyme (New England Biolabs, Ipswich, MA, USA) and electrophoresis was performed in a CHEF DR III system (Bio-Rad Laboratories, USA) using the following program: switch time 1–30s during 23h with a 120° angle at a temperature of 11.3°C and a voltage gradient of 6V/cm. The Lambda Ladder PFG marker kit (New England Biolabs, USA) was used as the DNA size marker. The gels were stained with ethidium bromide and digitally photographed using a Scorpion SCOR-14SOM scanner (DNR Bioimaging System, Jerusalém, Israel) under ultraviolet light. The images were analyzed using Gel ComparII® software (Applied Maths, Belgium). Electrophoretic profiles were compared according to the guidelines proposed by Tenover et al.24 The Dice coefficient (95%) and a 1% position tolerance were used to analyze the similarities in the band patterns among the electrophoretic profiles. The unweighted pair group method using the arithmetic average was used to gather electrophoretic profiles into polymorphism patterns, also referred to as clusters. The polymorphism patterns were defined grouping profiles that showed >70% dendrogram identity.
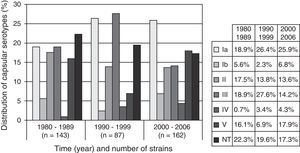
ResultsAnalyzing our serotyping data together with data previously described,3,16,25 the strains were classified as follows: Ia=23.5% (92 strains), Ib=5.3% (21), II=15.0% (59), III=18.9% (74), IV=2.8% (11) and V=14.8% (58). Serotype III prevailed among the human strains and type V among the bovine ones. Seventy-seven strains (19.7%) were non-typeable. Fig. 1 shows the distribution of serotypes along time.
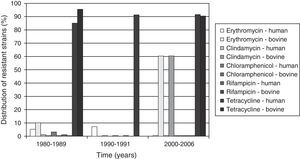
All the bovine and human isolates were susceptible to penicillin, vancomycin and levofloxacin. However, resistance to clindamycin (20.7% and 1.9% in bovine and human strains, respectively), erythromycin (27.6 and 4% in bovine and human strains, respectively), and tetracycline (89.6 and 89.2% in bovine and human strains, respectively) was detected among strains from both origins. Resistance to chloramphenicol and rifampicin was detected only among human originated strains, with rates of 1.5% and 0.7% respectively. All the erythromycin-resistant GBS strains from human origin were isolated from carriers or cases of noninvasive infections. All the clindamycin-resistant strains were also resistant to erythromycin. The distribution of resistant strains according to origin (bovine or human) and along time is shown in Fig. 2.
Constitutive macrolide-lincosamide-streptogramin B (cMLSB) and macrolide (M) phenotypes were equally distributed among the 23 erythromycin-resistant isolates, with 12 and 11 strains each one respectively, although the first prevailed among the bovine (75.0%) and the second among the human (60.0%) erythromycin-resistant GBS isolates. The inducible MLSB phenotype was not observed. The ermB gene was the most common resistance determinant in both human and bovine erythromycin-resistant isolates, while lnuB gene was not detected. Fourteen erythromycin-resistant strains analyzed (60.8%) had 2 or 3 erythromycin resistance determinants, and the combination ermB/mefA/E was predominantly found in 26.1% (6 strains). Only one S. agalactiae strain, isolated from a human host, did not harbor any of the erythromycin resistance genes tested. All M phenotype strains harbored the mefA/E gene, either alone or in combination with an erm (A or B) gene, while all the cMLSB phenotype strains had an erm gene, either alone or in combination with the mefA/E gene.
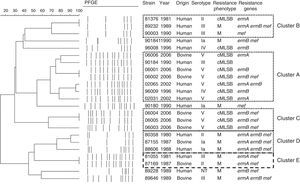
PFGE revealed a high heterogeneous population, as shown in Fig. 3, with 19 distinct electrophoretic profiles distributed among the 23 erythromycin-resistant GBS strains analyzed, suggesting a polyclonal origin for erythromycin resistance in our region. Four major clusters were observed, and the predominant one (cluster A) harbored 7 strains from both human and bovine origins. Similar genetic profiles were observed in different States of Brazil, as well as among strains belonging to different serotypes. In addition, one human originated serotype III strain from Rio de Janeiro and one bovine serotype II strain from São Paulo had identical profiles.
Genetic diversity and phenotypic characterization of S. agalactiae macrolide-resistant strains. Pulsed-field gel electrophoresis (PFGE) profiles, strain identification numbers, years of isolation, origins, serotypes, resistance phenotypes and resistance genes. cMLSB: constitutive macrolides, lincosamides and streptogramins B-type resistance; M, macrolides resistance; NT, non-typeable.
In United States, Europe and Australia serotypes Ia, II, III and V account for 80–90% of the isolated strains, while serotypes IV, VI, VII, VIII and IX remain rarely described.11,12,26,27 In the present study serotypes Ia, II, III and V were collectively detected in approximately 70% of the strains. This finding is consistent with previously reported data. Around 20% of our strains were non-typeable and similar data were described recently in our region.23 On the other hand, lower rates, varying between 4 and 15%, have been reported by others.8,28 The use of molecular techniques to detect the capsular gene in the latter may explain this discrepancy.
Susceptibility to vancomycin, penicillin and levofloxacin and a high level of resistance to tetracycline (89%) were observed in the present study. These data were also described previously in other countries13,29 and in Brazil.15,23,30,31 Although strains resistant to penicillin and levofloxacin have been described, they remain as exceptions.32,33
The tetracycline resistance rates can reach or overcome the 90% index. Tetracycline resistance rates observed among GBS isolates from humans agree with previous local and global studies.9,23,32 However, the rate among GBS from cattle is considerably higher than previous data, which had revealed only 44.7% and 14.5% of tetracycline resistance in Brazil30 and in the United States,9 respectively. The rise of tetracycline resistance is of concern since it is one of the main antimicrobials used in mastitis control.2,5 Moreover, according to our results, the general rate of tetracycline resistance (including strains from human and bovine origins) has increased from 86% to 91% from 80s to 90s and has been constantly high since then.
Although the rate of erythromycin resistance among S. agalactiae from human hosts analyzed in the present study has increased from the 80s to the 90s but decreased by 2006, it is still important to track erythromycin resistance among Brazilian S. agalactiae strains. Recently, rates around 13% and 11%, which would rule out this drug as an alternative option, have been described in Rio de Janeiro.23,33 Among strains from bovine origin, resistance to erythromycin increased from 10.5% in 1987 and 1988 to 60% in 2003 and 2006. This is probably also due to the proximity among the collection herds investigated in 2006; hereafter mentioned.
Prevalence of cMLSb phenotype increased along the years, while prevalence of M phenotype decreased. The predominance of MLSb phenotype among S. agalactiae strains obtained in the last ten years was reported in Brazil and in other countries.23,33–35 The predominant resistance genotype was ermB+ mefA/E+, suggesting that the majority of our strains have both resistance mechanisms, efflux and ribosomal modification. The three states included in the study (Rio de Janeiro, Santa Catarina, and São Paulo) were represented among the 23 erythromycin-resistant GBS, and no correlation between erythromycin resistance and a specific serotype was observed, since resistant strains belonged to a variety of them (Ia, II, III, IV and V, and NT).
The increasing resistance to macrolides among group B streptococci has been observed during recent years in many countries. It is particularly important from the epidemiological point of view to perform specific characterization of S. agalactiae isolates, focused on, among others, answering if the phenomenon occurred due to the spread of a specific S. agalactiae clone in the population or if it is the result of acquired resistance or both. Some authors have associated macrolide resistance to the introduction of specific S. agalactiae clones, including mostly serotype V strains.28,34,36 The present study, however, did not show association between specific S. agalactiae clones or serotypes and the macrolide resistance profile among strains from human origin, indicating that horizontal genetic transfer is the main force behind the appearance of erythromycin resistance in our region. Similar data were described previously for S. agalactiae strains isolated in 2008 in Rio de Janeiro.23 Indeed, some authors have shown that erythromycin resistance determinants are carried by conjugative transposons, which are easily self-transferable and commonly found among S. agalactiae strains.37,38
Regarding the bovine strains, however, it is important to highlight the serotype V cluster (cluster C) observed in 2006. These isolates were recovered from herds in the South Central region of Rio de Janeiro and, although more studies are necessary to evaluate this hypothesis, it seems that they represent a successful clone, well-adapted to the local cattle, and dissemination to the metropolitan region, which is nearby, could easily happen. Moreover, the finding of a major cluster, cluster A, comprising human and bovine originated GBS strains suggests that, although erythromycin resistance is rather emerging due to horizontal genetic transfer, successful clones, which can be found among strains from humans and cattle, have the ability to capture and exchange this kind of mobile genetic elements.
A GBS strain from human origin, isolated in Rio de Janeiro in 1981, and one from cattle, isolated in São Paulo in 1987, showed indistinguishable electrophoretic profiles (cluster E). The genetic relationship between bovine and human originated strains has been for a long time speculated and previous studies around the world have, as well, demonstrated very similar or identical genotypes among them.39–41 In fact, we ourselves have earlier reported human and bovine GBS strains sharing genetic (same PFGE profile, ribotype and related ST) and phenotypic (in vivo virulence profile) characteristics, indicating a common evolutionary origin and suggesting that strains from different hosts could presumably transfer genetic material between each other.25,42 Nevertheless, there is still a lot to elucidate about this hypothesis.
ConclusionIn the present study, almost four hundred S. agalactiae human and bovine strains collected during a 27-year period were characterized by phenotypic and genetic aspects, generating original and valuable data on serological typing, antimicrobial susceptibility profile and genetic diversity, in order to help elucidating GBS epidemiology in Brazil. Furthermore, our data about macrolide resistant strains, a major concern regarding the epidemiology of S. agalactiae, point out that it is rather due to horizontal gene transfer than to monoclonal dissemination among Brazilian strains.
Conflict of interestAll authors declare to have no conflict of interest.
This work was supported by CNPq, FAPERJ, CAPES, PRONEX, and The Thrasher Research Fund.