The emergence of antibiotic resistance is increasing and there are few effective antibiotics to treat infections caused by resistant and multidrug resistant bacterial pathogens. This study aimed to evaluate the in vitro activity of ceftolozane–tazobactam against clinical bacterial isolates from Brazil.
MethodsA total of 673 Gram-negative bacterial isolates including Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa and other Enterobacterales collected from 2016 to 2017 were tested, most of them isolated from patients in intensive care units. Minimum inhibitory concentrations (MIC50/90) were determined by broth microdilution for amikacin, aztreonam, cefepime, cefotaxime, cefoxitin, ceftolozane–tazobactam, ceftazidime, ceftriaxone, ciprofloxacin, colistin, ertapenem, imipenem, levofloxacin, meropenem, and piperacillin-tazobactam using dried panels. Antimicrobial susceptibility results were interpreted according to Clinical and Laboratory Standards Institute criteria.
ResultsSusceptibility rates to ceftolozane–tazobactam ranged from 40.4% to 94.9%. P. aeruginosa susceptibility rate to ceftolozane–tazobactam was 84.9% (MIC50/90, 1/16μg/mL) and 99.2% to colistin. For E. coli, ceftolozane–tazobactam inhibited 94.9% (MIC50/90, 0.25/1μg/mL) of the microorganisms. The susceptibility rate of K. pneumoniae to ceftolozane–tazobactam was 40.4% (MIC50/90, 16/>32μg/mL). Other Enterobacterales have shown susceptibility rates of 81.1% (MIC50/90, 0.5/16μg/mL) to ceftolozane–tazobactam, 93.9% to meropenem, 90.9% to amikacin (90.9%), and 88.6% to ertapenem. In non-carbapenemase producing isolates, AmpC mutations were found three isolates.
ConclusionsCeftolozane–tazobactam has shown relevant activity against a large variety of the analyzed microorganisms collected from multiple centers in Brazil, showing promising results even in multidrug resistant strains.
The emergence of antibiotic resistance is increasing and represents one of the major threats to global health today. Infections caused by multidrug resistant microorganisms cause relevant clinical and economic impact leading to increased hospital stays, costs and mortality.1 The Centers for Disease Control and Prevention (CDC) points that about two million people in the USA present with infections caused by bacteria resistant to at least one antimicrobial agent of choice each year, leading to at least 23,000 deaths annually.2 Healthcare-associated infections account for a high incidence of multidrug resistant microorganisms due to recurrent overuse and misuse of these medications, high density of susceptible hosts, and increased odds of transmission among patients.3,4
In 2017, the World Health Organization (WHO) published a list of pathogens with priority demand for new antibiotics due to antimicrobial resistant profile. Amongst the critical priority pathogens were carbapenem-resistant Pseudomonas aeruginosa, and carbapenem and third-generation cephalosporins-resistant Enterobacterales, which includes Escherichia coli, Klebsiella pneumoniae, Enterobacter spp., Serratia spp., Proteus spp., Providencia spp., and Morganella spp.5
P. aeruginosa has a leading role among infections caused by Gram-negative rods, especially in critically ill and immunocompromised patients, e.g. those who experienced extensive, high-degree burns.6 Treatment options are limited due to antimicrobial resistance, which permitted P. aeruginosa infections to become a serious issue causing a total of 51,000 healthcare-associated infections per year in the USA.7,8 In Brazil, P. aeruginosa is the fifth bacterial pathogen in bloodstream infections associated with central venous catheters in adult patients, accounting for 8.5% of this infection in 2016.9
Currently, eight categories of antibiotics are mainly used to treat P. aeruginosa infections such as aminoglycosides, carbapenems, cephalosporins, fluoroquinolones, penicillin with β-lactamase inhibitors, monobactams, fosfomycin, and polymyxin. However, Gram-negative bacilli like P. aeruginosa may present many mechanisms of antibiotic resistance, which can be expressed at the same time, including decrease in influx porin activity, increase in efflux pumps, production of altered penicillin-binding proteins and catalytic enzymes that inactivate antibiotics.7,10
Meanwhile, Enterobacterales also represent a significant concern due to carbapenem-resistance.11 Currently, therapies against carbapenem-resistant Enterobacterales include tigecycline, ceftazidime-avibactam, and other drugs not widely used recently such as colistin, fosfomycin, and aminoglycosides.11
Few studies have reported the profile of new antibiotics in low-to middle income countries. Pfaller et al. reported the in vitro activity of ceftolozane–tazobactam and comparator agents against Latin American isolates Enterobacterales and P. aeruginosa from 2013 to 2015.12 Considering the need for updated knowledge about susceptibility profile, especially regarding new antibiotics, this study aimed to evaluate the in vitro activity of ceftolozane–tazobactam against clinical bacterial isolates of P. aeruginosa, E. coli, K. pneumoniae, and other Enterobacterales from Brazil.
Materials and methodsStudy designThis study evaluated the antibacterial activity and minimum inhibitory concentration of ceftolozane–tazobactam and 15 comparator compounds against a recent collection of Gram-negative bacterial isolates prospectively collected in 10 different centers in Brazil from 2016 to 2017. Centers were in the following Brazilian states: Bahia, Minas Gerais, Paraná, Rio de Janeiro, and São Paulo.
Bacterial isolatesThe bacteria used in the analysis were selected from different participating sites across the country. Each site collected consecutive fresh clinical Gram-negative aerobic isolates composed only by: E. coli, K. pneumoniae, P. aeruginosa, and other Enterobacterales (Proteus mirabilis, Enterobacter cloacae, and K. oxytoca). No more than 15% of the isolates ought to be retrieved from lower respiratory tract infections. The remainder were to be collected from complicated urinary infections and complicated intra-abdominal infections sources. Only isolates associated with infection were included in the study. Multiple organisms from one specimen were acceptable provided each was a unique initial Gram-negative bacillus, but only the first isolate of a particular species per patient for the entire collection period was accepted. Exclusion criteria were: non-target species; isolates from in situ drains or drainage bottles; isolates from stools or perirectal abscess; duplicate isolates (same genus and species) obtained from the same or different specimen even if they were obtained from the same or different body sites; duplicate isolates (same genus and species) obtained at any subsequent time from the same patient, regardless of susceptibility or phenotypic profile; and environmental samples (non-patient derived) or surveillance cultures taken for infection control purposes.
After initial identification of the isolates in the primary center, all isolates were stored in Tryptic Soy Broth with glycerol and frozen at −70 or −20°C. All isolates were further sent for a reference laboratory (International Health Management Associates Inc.). All isolates were re-identified by the coordinator center using MALDI-TOF MS.
Minimum inhibitory concentrationMIC values were determined by broth microdilution according to CLSI guidelines for antimicrobials previously described, using dried panels prepared by Trek Diagnostics Systems Ltd. (East Grinstead, West Sussex, UK).13,14 Ceftolozane–tazobactam, amikacin, aztreonam, cefepime, cefotaxime, cefoxitin, ceftazidime, ceftriaxone, ciprofloxacin, colistin, ertapenem, imipenem, levofloxacin, and piperacillin-tazobactam were determined using broth microdilution as shown in Table 1.
Range and concentrations of compounds tested.
| Compound | Test range (mg/L) |
|---|---|
| Amikacin | 4–32 |
| Aztreonam | 1–16 |
| Cefepime | 1–32 |
| Cefotaxime | 1–32 |
| Cefoxitin | 2–16 |
| Ceftolozane–tazobactam | 0.06–32 |
| Ceftazidime | 1–32 |
| Ceftriaxone | 1–32 |
| Ciprofloxacin | 0.25–2 |
| Colistin | 1–4 |
| Ertapenem | 0.06–4 |
| Imipenem | 0.5–32 |
| Levofloxacin | 1–4 |
| Meropenem | 0.12–16 |
| Piperacillin-tazobactam | 2–64 |
Colonies were taken directly from a second-pass culture plate and prepared to a suspension equivalent of the 0.5 McFarland standard using normal saline. Inoculation of the MIC plates took place within 15min after adjustment of the inoculum suspension turbidity. The panels were incubated at 35°C for 16–20h before reading the MIC endpoints. Interpretive criteria followed CLSI 2017 guidelines for all compounds except colistin for which the EUCAST 2017 breakpoint was used for Enterobacterales.14,15 Quality control (QC) testing was performed each day of testing as specified by the CLSI using E. coli ATCC 25922, P. aeruginosa ATCC 27853, and K. pneumoniae ATCC 700603. All QC data were within CLSI approved ranges where available.14
Presence of carbapenemases in C/T resistant P. aeruginosaPhenotypic detection of carbapenemases was performed by the Carba-NP (Repidec®) test, following manufacturer's protocol on all C/T non-susceptible isolates (MIC50/90 ≥8/4).
Molecular detection of carbapenemases was performed on all Carba-NP positive isolates by in-house conventional PCR assays, which targeted genes encoding the following groups of carbapenemases: KPC, NDM, VIM, IMP, and OXA-48.
Whole genome sequencing of resistant strainsGenomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). DNA quality was verified by agarose gel electrophoresis and quantified using Qubit 2.0 fluorometer (Invitrogen by Thermo Fisher Scientific, Life Technologies Italia, Monza, Italy). Paired-end libraries were prepared from 1ng of total bacterial DNA using Nextera XT DNA Sample Preparation kit and Nextera XT Index kit (Illumina Inc., San Diego, California, U.S.A.). Library concentration and average fragment size were calculated by Qubit 2.0 fluorometer and Agilent 2100 bioanalyzer (Agilent technologies, Santa Clara, CA, U.S.A.). Sequences were obtained on the Illumina MiSeq platform (Illumina Inc., San Diego, California, U.S.A.) with 250nt paired-end reads to achieve a coverage of about 30× per base, using MiSeq V3 flow cell.
Raw reads underwent a series of steps for quality filtering which included a general quality check and the trimming of low-quality ends. De novo assemblies were performed using CLC Genomics Workbench 8.1.0 software. Annotation was done by using RAST server, and the presence of resistance determinants were determined by BLAST and ResFinder.
The genome of P. aeruginosa PAO1 (GenBank ID: NC_002516.2 https://www.ncbi.nlm.nih.gov/genome/187?genome_assembly_id=299953) was used as reference to look for known substitutions associated with C/T resistance in AmpC and its regulator, AmpR.
Statistical analysisData were analyzed using a descriptive approach through measures of central tendency and dispersion for continuous variables and measures of frequency for categorical variables. MICs50/90 were calculated.
ResultsA total of 673 isolates from 10 sites were included during the study period. The clinical isolates were retrieved from intensive care units (ICU) (75%) and from non-ICU (25%).
Table 1 shows the antibiotics assessed, the range and concentration of each drug, and Table 2 shows the distribution of species tested. E. coli (N=216; 32.1%), K. pneumoniae (N=193; 28.7%), and P. aeruginosa (N=132; 19.6%) were the most prevalent species.
Distribution of included isolates by species in Brazil.
| Species | N | Percent (%) |
|---|---|---|
| Escherichia coli | 216 | 32.1 |
| Klebsiella pneumoniae | 193 | 28.7 |
| Pseudomonas aeruginosa | 132 | 19.6 |
| Proteus mirabilis | 72 | 10.7 |
| Enterobacter cloacae | 38 | 5.6 |
| Klebsiella oxytoca | 11 | 1.6 |
| Klebsiella variicola | 4 | 0.6 |
| Klebsiella aerogenes | 3 | 0.4 |
| Enterobacter asburiae | 1 | 0.1 |
| Morganella morganii | 1 | 0.1 |
| Proteus vulgaris | 1 | 0.1 |
| Providencia rettgeri | 1 | 0.1 |
| Total | 673 | 100 |
Table 3 shows the susceptibility profile of different organisms to several antibiotics.
In vitro activity of ceftolozane–tazobactam and comparators against 673 isolates from Brazil.
| Organism (n) | Antimicrobial | %S | %I | %R | MIC 50 | MIC90 | Range |
|---|---|---|---|---|---|---|---|
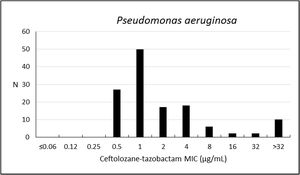
| Pseudomonas aeruginosa (132) | Ceftolozane–tazobactam | 84.9 | 4.6 | 10.6 | 1 | 16 | 0.5–>32 |
| Amikacin | 84.9 | 3.0 | 12.1 | ≤4 | >32 | ≤4–>32 | |
| Aztreonam | 50.0 | 15.9 | 34.1 | 8 | >16 | 2–>16 | |
| Cefepime | 61.4 | 12.9 | 25.8 | 8 | >32 | ≤1–>32 | |
| Ceftazidime | 59.9 | 9.9 | 30.3 | 8 | >32 | ≤1–>32 | |
| Ciprofloxacin | 68.9 | 6.8 | 24.2 | ≤0.25 | >2 | ≤0.25–>2 | |
| Colistin | 99.2 | – | 0.8 | ≤1 | ≤1 | ≤1–4 | |
| Imipenem | 53.0 | 6.1 | 40.9 | 2 | 32 | ≤0.5–>32 | |
| Levofloxacin | 62.9 | 7.6 | 29.6 | ≤1 | >4 | ≤1–>4 | |
| Meropenem | 58.3 | 9.9 | 31.8 | 2 | >16 | ≤0.12–>16 | |
| Piperacillin-tazobactam | 55.3 | 14.4 | 30.3 | 16 | >64 | ≤2–>64 | |
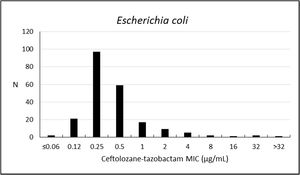
| Escherichia coli (216) | Ceftolozane–tazobactam | 94.9 | 2.3 | 2.8 | 0.25 | 1 | ≤0.06–>32 |
| Amikacin | 97.7 | 0.9 | 1.4 | ≤4 | 8 | ≤4–>32 | |
| Aztreonam | 67.1 | 7.4 | 25.5 | ≤1 | >16 | ≤1–>16 | |
| Cefepime | 65.3 | 7.9 | 26.9 | ≤1 | >32 | ≤1–>32 | |
| Cefotaxime | 60.7 | 1.4 | 38.0 | ≤1 | >32 | ≤1–>32 | |
| Cefoxitin | 68.5 | 14.8 | 16.7 | 8 | >16 | ≤2–>16 | |
| Ceftazidime | 70.4 | 9.7 | 19.9 | ≤1 | 16 | ≤1–>32 | |
| Ceftriaxone | 59.7 | 1.4 | 38.9 | ≤1 | >32 | ≤1–>32 | |
| Ciprofloxacin | 47.7 | 1.9 | 50.5 | >2 | >2 | ≤0.25–>2 | |
| Colistin | 98.2 | – | 1.9 | ≤1 | ≤1 | ≤1–4 | |
| Ertapenem | 94.0 | 0.5 | 5.6 | ≤0.06 | 0.12 | ≤0.06–>4 | |
| Imipenem | 97.2 | 1.9 | 0.9 | ≤0.5 | ≤0.5 | ≤0.5–8 | |
| Levofloxacin | 49.5 | 3.7 | 46.8 | 4 | >4 | ≤1–>4 | |
| Meropenem | 97.2 | 1.4 | 1.4 | ≤0.12 | ≤0.12 | ≤0.12–>16 | |
| Piperacillin-tazobactam | 91.7 | 3.7 | 4.6 | ≤2 | 16 | ≤2–>64 | |
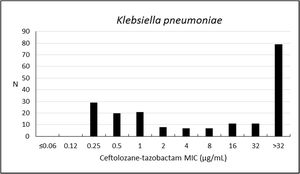
| Klebsiella pneumoniae (193) | Ceftolozane–tazobactam | 40.4 | 3.6 | 56.0 | 16 | >32 | 0.25–>32 |
| Amikacin | 91.2 | 1.6 | 7.3 | ≤4 | 16 | ≤4–>32 | |
| Aztreonam | 23.8 | 3.1 | 73.1 | >16 | >16 | ≤1–>16 | |
| Cefepime | 25.4 | 7.8 | 66.8 | >32 | >32 | ≤1–>32 | |
| Cefotaxime | 21.8 | 2.1 | 76.2 | >32 | >32 | ≤1–>32 | |
| Cefoxitin | 39.9 | 10.4 | 49.7 | 16 | >16 | ≤2–>16 | |
| Ceftazidime | 28.5 | 2.1 | 69.4 | >32 | >32 | ≤1–>32 | |
| Ceftriaxone | 20.7 | 1.6 | 77.7 | >32 | >32 | ≤1–>32 | |
| Ciprofloxacin | 25.9 | 2.1 | 72.0 | >2 | >2 | ≤0.25–>2 | |
| Colistin | 90.2 | – | 9.8 | ≤1 | 2 | ≤1–>4 | |
| Ertapenem | 53.4 | 1.0 | 45.6 | 0.5 | >4 | ≤0.06–>4 | |
| Imipenem | 61.1 | 4.7 | 34.2 | ≤0.5 | 32 | ≤0.5–>32 | |
| Levofloxacin | 28.5 | 4.7 | 66.8 | >4 | >4 | ≤1–>4 | |
| Meropenem | 60.1 | 2.1 | 37.8 | ≤0.12 | >16 | ≤0.12–>16 | |
| Piperacillin-tazobactam | 30.6 | 14.0 | 55.4 | >64 | >64 | ≤2–>64 | |
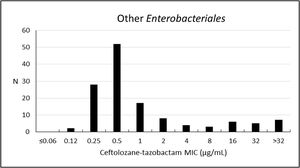
| Other Enterobacteralesa (132) | Ceftolozane–tazobactam | 81.1 | 3.0 | 15.9 | 0.5 | 16 | 0.12–>32 |
| Amikacin | 90.9 | 2.3 | 6.8 | ≤4 | 16 | ≤4–>32 | |
| Aztreonam | 72.0 | 0.8 | 27.3 | ≤1 | >16 | ≤1–>16 | |
| Cefepime | 62.1 | 10.6 | 27.3 | ≤1 | >32 | ≤1–>32 | |
| Cefotaxime | 52.3 | 3.0 | 44.7 | ≤1 | >32 | ≤1–>32 | |
| Cefoxitin | 56.1 | 6.1 | 37.9 | 8 | >16 | ≤2–>16 | |
| Ceftazidime | 72.7 | 2.3 | 25.0 | ≤1 | >32 | ≤1–>32 | |
| Ceftriaxone | 52.3 | 5.3 | 42.4 | ≤1 | >32 | ≤1–>32 | |
| Ciprofloxacin | 63.6 | 2.3 | 34.1 | ≤0.25 | >2 | ≤0.25–>2 | |
| Colistin | 40.9 | – | 59.1 | >4 | >4 | ≤1–>4 | |
| Ertapenem | 88.6 | 3.8 | 7.6 | ≤0.06 | 1 | ≤0.06–>4 | |
| Imipenem | 72.0 | 18.9 | 9.1 | ≤0.5 | 2 | ≤0.5–>32 | |
| Levofloxacin | 68.9 | 4.6 | 26.5 | ≤1 | >4 | ≤1–>4 | |
| Meropenem | 93.9 | 0 | 6.1 | ≤0.12 | 0.25 | ≤0.12–>16 | |
| Piperacillin-tazobactam | 80.3 | 7.6 | 12.1 | ≤2 | >64 | ≤2–>64 |
%S, I, R: percent susceptible, intermediate, resistant by CLSI 2017 guidelines, EUCAST guidelines for colistin were applied for Enterobacterales; MIC50, MIC90, and range in μg/mL; –, no intermediate breakpoint.
Considering the analysis performed for P. aeruginosa, the susceptibility rate to ceftolozane–tazobactam was 84.9%, with MIC50/90 values of 1/16μg/mL (Table 3; Fig. 1). Colistin was the most active compound, with 99.2% of isolates susceptible (MIC50/90, ≤1/≤1μg/mL); cefepime, ciprofloxacin, and levofloxacin have also shown estimates above 60%.
Ceftolozane–tazobactam susceptibility rate against E. coli was 94.9%. Susceptibility rates to other antibiotics were 97.7% to amikacin, 97.2% to meropenem, 94.0% to ertapenem, and 91.7% to piperacillin-tazobactam. The activity of ceftolozane–tazobactam against E. coli was high (MIC50/90, 0.25/1μg/mL) (Table 3; Fig. 2).
Susceptibility rate of K. pneumoniae to ceftolozane–tazobactam was 40.4%, with weak activity against this microorganism (MIC50/90, 16/>32μg/mL), but higher susceptibility to amikacin (91.2%), colistin (90.2%), imipenem (61.1%), meropenem (60.1%), and ertapenem (53.4%) (Table 3; Fig. 3). Other antibiotics have shown inhibition frequencies that ranged from 20.7% to 39.9% (Table 3; Fig. 3).
The analysis performed with other Enterobacterales have shown a susceptibility rate to ceftolozane–tazobactam of 81.1% (MIC50/90, 0.5/16μg/mL). These microorganisms had greater susceptibility to meropenem (93.9%), amikacin (90.9%), and ertapenem (88.6%) – Table 3; Fig. 4.
Frequency distribution (n) of ceftolozane–tazobactam at each MIC (μg/mL) for 132 other* Enterobacterales from Brazil.
* Other Enterobacterales consist of (n): Klebsiella aerogenes (3); E. asburiae (1); E. cloacae (38); Klebsiella oxytoca (11); K. variicola (4); Morganella morganii (1); Proteus mirabilis (72); P. vulgaris (1); Providencia rettgeri (1).
From 13 ceftolozane/tazobactam non-susceptible P. aeruginosa isolates, carbapenemase was detected in 8/13 (62%) isolates which fully explains the detected resistance. The presence of at least one carbapenemase gene was demonstrated in only one isolate (KPC), while the identity of the carbapenemase gene harbored by the remaining 7/8 strains (87.5%) remains unknown and is yet to be studied.
From the remaining five (38%) isolates tested negative for the presence of carbapenemases by Carba-NP, three were randomly chosen to be sequenced aiming to detect previously reported mutations associated to the ceftolozane/tazobactam resistant phenotype.
As expected, genes encoding aminoglycoside modifying enzymes (aph(3′β-IIb)), chloramphenicol acetyltransferases (catB7), and fosfomycin resistance enzymes (fosA) were found in all three isolates. Regarding β-lactamases genes, blaPAO (blaAmpC) and blaOXA-50 were also detected in all isolates tested.
AmpC substitutions G27D, V205L, and G391A were all found in only one isolate, while substitution T105A was found in two isolates. On the other hand, AmpC substitutions E114A, G283E, and M288R were all detected in only one is olate.
DiscussionThis study was conducted aiming to evaluate the activity of ceftolozane–tazobactam against clinical bacterial isolates from Brazil. Thus, in vitro analyses were performed and susceptibility of different organisms to several antibiotics are presented.
Microorganisms tested have shown susceptibility rates to ceftolozane–tazobactam ranging from 40.4% (K. pneumoniae) to 94.9% (E. coli). The susceptibility profile to ceftolozane–tazobactam have varied according with microorganism by intrinsic and extrinsic mechanisms and also according to the country that provided the samples.12,16–27 Shortridge et al. assessed the activity of ceftolozane–tazobactam against Gram-negative isolates from bloodstream infection in the US from 2013 to 2015. In that study, E. coli NDM-1, K. pneumoniae NDM-1, and P. aeruginosa pan-β-lactams non-susceptible (pan-BL-NS) presented complete resistance. P. aeruginosa pan-BL-NS were non-susceptible to all β-lactams including ceftolozane–tazobactam. Isolates containing carbapenemases or metallo-beta-lactamases were uncommon (1.4%) in the study, which compromised the robustness of the findings.25 However, three studies have reported susceptibility to ceftolozane–tazobactam that reached 100%, against the following microorganisms: E. coli CTX-M 14-like, E. coli SHV ESBL, E. coli TEM ESBL, K. pneumoniae CTX-M 14-like, Citrobacter koseri, Proteus mirabilis and Proteus spp.23,25,28 Garcia-Fernandez et al. highlight the activity of ceftolozane–tazobactam against wild type Enterobacterales, although it is dependent on both the species and resistance mechanisms.28 In our study, the susceptibility of Klebsiella spp. to ceftolozane–tazobactam was low. This was due to high incidence of carbapenemases in this microorganism, most of them with high rate of hydrolysis of ceftolozane, like KPC. This result suggests that ceftolozane–tazobactam should be used cautiously in empirical therapies considering the high incidence of KPC-producing bacteria. It is important to note that MIC cut-off for Enterobacterales was 1mg/L, compared to 4mg/L for P. aeruginosa. This is due to higher affinity of ceftolozane to P. aeruginosa binding sites.
Regarding the observed susceptibility of P. aeruginosa, 84.9% were susceptible to ceftolozane–tazobactam. The same pattern was observed for other Enterobacterales, with susceptibility rates above 80%. These microorganisms represent a great concern for clinical management on community and hospital acquired infections due to its resistance to available antibiotics. Carbapenem resistance observed in Enterobacterales, usually mediated by transferable β-lactamase enzymes, has also become an important issue.29
Some authors have studied the activity of ceftolozane–tazobactam specifically against P. aeruginosa and Enterobacterales isolates from Germany, USA, Spain, and Latin America. The frequency of susceptibility rates ranged from 30% to 100% for P. aeruginosa and from 77.8% to 100% for Enterobacterales.12,18,20,23–26,28 Pfaller et al. reported results of ceftolozane–tazobactam activity against multidrug-resistant Enterobacterales and P. aeruginosa from Latin America, including isolates from Brazil. Ceftolozane–tazobactam was the most active of the β-lactam agents tested against P. aeruginosa and it was the second most active drug, after meropenem, against Enterobacterales.12
Garcia-Fernandez et al. reported the activity of ceftolozane–tazobactam specifically in the context of intensive care units, where a worse pattern of resistance is observed. When P. aeruginosa was assessed, a susceptibility rate of 91.3% was observed and it was the third most active, after colistin (95.0%) and amikacin (93.8%). When Enterobacterales were assessed, ceftolozane–tazobactam activity ranged from 5% (carbapenemase-phenotype K. pneumoniae) to 100% (Proteus spp.),28 which is aligned with the most recent national surveillance findings, where almost 45% of K. pneumoniae bloodstream infection isolates were resistant to 3rd and 4th generation cephalosporins and carbapenems.30
Some resistant isolates produce enzymes able to degrade or inactivate β-lactams, such as ESBL samples.31 Being a combination of a β-lactam and an inhibitor of a considerable number of β-lactamase enzymes, ceftolozane–tazobactam retains activity against isolates resistant to other β-lactams, sparing the use of meropenem.32–34 It is well known that carbapenem use results in escalating rates of carbapenem-resistant infections, limiting treatment options and increasing mortality.35
In this context, a sub-analysis was performed in order to describe genetic profile of resistant mechanisms in ceftolozane/tazobactam non-susceptible P. aeruginosa isolates. The presence of a carbapenemase in most of the isolates is consistent with results previously described in the country.36–39 Although one isolate had at least one carbapenemase gene, the remaining still needs further analysis to better address this issue. In Brazil, it is estimated that the frequency of carbapenem-resistant P. aeruginosa may reach 40.0% of the observed infections.39–42 The results shown in the present study highlights the presence of intrinsic mechanisms, and that not all cases of resistance were mediated by transferable carbapenemase-encoding genes. Ceftolozane–tazobactam resistance in non-carbapenemase producing isolates were associated with ampC over-expression by single point mutations in the blaPDC gene (ampC gene) identified by whole gene sequencing. AmpC substitutions G27D, V205L, G391A, T105A, E114A, G283E, and M288R were all associated with AmpC overexpression.
When compared to polymyxins and aminoglycosides, ceftolozane–tazobactam has a greater safety profile.1 Polymyxins and aminoglycosides are antibiotics with high frequency of toxicity and the last choice when modern antimicrobials are not effective or contraindicated1,43 In a meta-analysis, colistin showed a similar safety profile when compared with other antibiotics in patients with ventilator-associated pneumonia.44 However, numerous adverse events, such as mortality, nephrotoxicity, neurotoxicity, allergic and topical reactions, were related with the use of polymyxins, highlighting its toxicity.43 Thus, since ceftolozane–tazobactam has a favorable safety profile, it seems to be a good option in clinical practice.
One study limitation needs to be highlighted: the lack of an alternative test to confirm MIC results of the isolates susceptibility profile. Despite MIC testing recommendation for antimicrobial susceptibility assessment, the study would benefit from using an additional technique or different approach to further investigate that point. Even though, the study provides new information about the activity of this antimicrobial agent against different isolates from Brazil.
As a regional surveillance network that collected 2410 pathogens from selected medical centers, the study does not provide a population-based data, nor information about the incidence of infections in a given region.
In conclusion, ceftolozane–tazobactam has shown satisfactory activity against a great variety of microorganisms analyzed, especially when respiratory tract is the infection site as reported in clinical studies, showing promising results even against resistant strains.17,21,45,46
The activity of ceftolozane–tazobactam is specially observed against P. aeruginosa, E. coli, and other Enterobacterales. Our data suggest that ceftolozane–tazobactam may be an important treatment option, including against multidrug resistant strains.
Editorial supportMedical writing was provided by Roberta Arinelli Fernandes, Ana Carolina Padula Ribeiro Pereira and Ayke Adnet de Lima of Sense Company Brazil. This assistance was funded by MSD Brazil.
Authors contributionAll authors were involved in at least one of the following: conception, design of work or acquisition, analysis, interpretation of data and drafting the manuscript and/or revising/reviewing the manuscript for important intellectual content. All authors provided final approval of the version to be published. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflicts of interestSuellen da Silva Rodrigues, Fernando Brandão Serra, and Marina Della-Negra de Paula, are employees of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ USA, who may own stock and/or hold stock options in Merck & Co., Inc., Kenilworth, NJ, USA.
FFT received research grants from MSD, Pfizer and TEVA.
AcknowledgmentThis study was supported by Merck Sharpe Dohme.