The emergence of KPC-2 producing K. pneumoniae in hospitalized patients at the intensive care unit (ICU) of a teaching hospital located in the city of João Pessoa, Paraíba, Brazil, is reported. Seven carbapenem-resistant K. pneumoniae recovered from different body sites of infection were analyzed. Most isolates showed a multidrug-resistance phenotype. Genotypic analysis demonstrated the presence of two genotypes, with the predominance of genotype A, which belongs to ST 437. These isolates also carry the encoding genes of five other beta-lactamases.
Klebsiella pneumoniae carbapenemases (KPC)-producing Enterobacteriaceae isolates have become a significant problem worldwide. KPC are enzymes that hydrolyze penicillins, cephalosporins, monobactams, and carbapenems, and are inhibited by clavulanic acid and tazobactan.1 Of notice, genes encoding KPC enzymes are frequently located on transferable plasmids. KPC-producing clinical isolates have been associated with hospital outbreaks in the USA, Asia, Europe, South Korea, and Central and South America.2 In Brazil, KPC-2-producing isolates have been initially reported in the cities of Recife, Rio de Janeiro, São Paulo, and Porto Alegre.3–9 In this report, the emergence of KPC-producing-K. pneumoniae (KPC-KPN) clinical isolates recovered at the tertiary teaching Hospital Lauro Wanderley, located in João Pessoa, Paraíba, Brazil is described.
In the period between 2009 and 2010, seven K. pneumoniae isolates were collected from different body sites of infection of five patients who were hospitalized at the intensive care unit (ICU) of the Hospital Lauro Wanderley. Initially, these isolates were identified as resistant to ertapenem by disc diffusion method, and were referred to the Laboratório Alerta, UNIFESP, São Paulo, Brazil for further characterization. Susceptibility testing was performed by the agar dilution method, according to the Clinical and Laboratory Standards Institute's (CLSI) guidelines10 for amikacin, aztreonam, cefepime, cefoxitin, ceftazidime, ceftriaxone, ciprofloxacin, ertapenem, imipenem, meropenem, and piperacillin/tazobactam. Minimum inhibitory concentrations (MICs) for tigecycline and polymyxin B were determined by Etest (bioMérieux – Marcy l’Etóile, France) and interpreted according to the European Committee on Antimicrobial Susceptibility Testing's (EUCAST) guidelines.11 The Modified Hodge Test (MHT) with ertapenem disk (10μg) was used for phenotypic detection of carbapenemase activity.12 Specific primers were used under standard polymerase chain reaction (PCR) conditions to detect the genes blaKPC, blaGES, blaCTX-M, blaSHV, blaTEM, blaOXA-1, blaOXA-2, blaOXA-10, blaOXA-18/45, and blaOXA-161, followed by DNA sequencing (ABI sequencer - Applied Biosystems, Foster City, CA). Clonal relatedness among isolates was examined by pulsed field gel electrophoresis (PFGE) using SpeI (New England, Beverly, MA). The band patterns were analyzed by visual interpretation, applying the criteria established by Tenover et al.13 In addition, the PFGE patterns were analyzed using Bionumerics version 5.10 (Applied Maths, Sint-Martens – Latem, Belgium) for comparison with other KPC-2-producing strains previously identified in Brazil.3 Multilocus sequence typing (MLST) of K. pneumoniae was performed as described previously.14 Experimentally determined DNA sequences were uploaded into the MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html), and allelic numbers and sequence types (ST) were obtained.
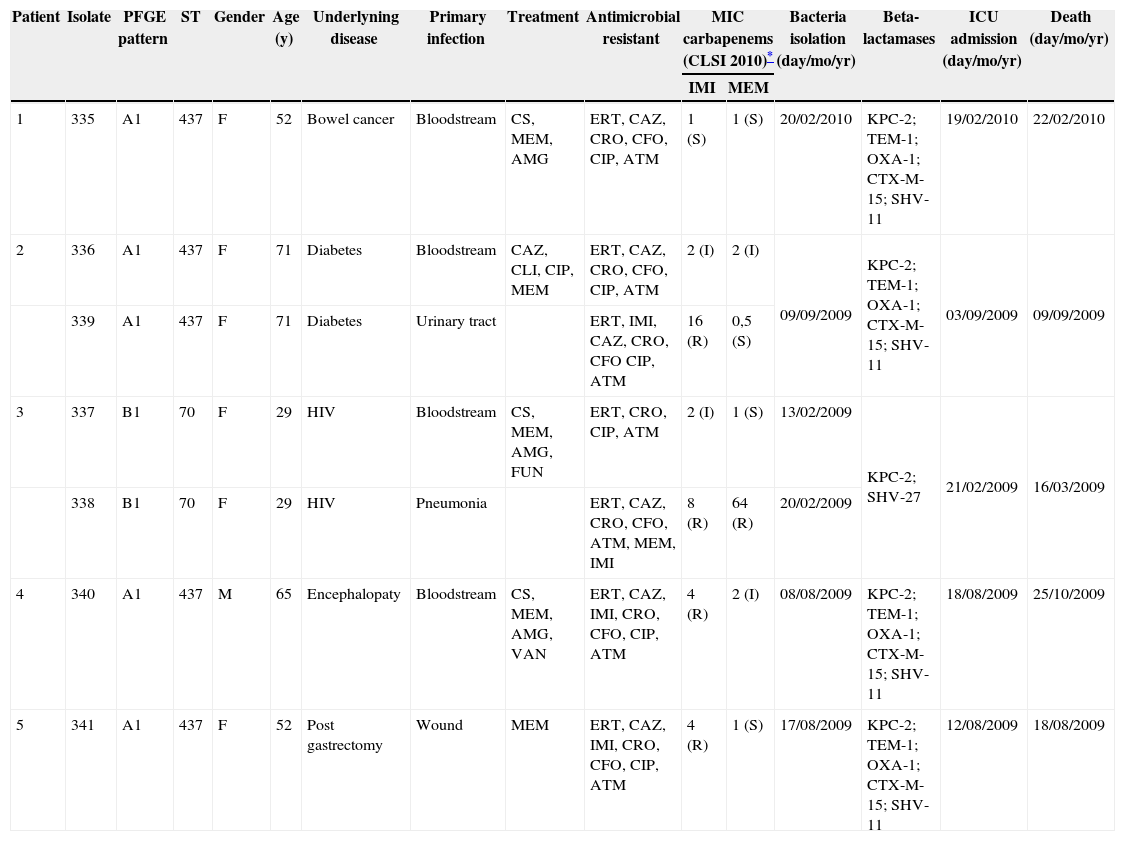
K. pneumoniae isolates were more frequently collected from bloodstream infections (n = 4; 57.1%) (Table 1). Amikacin, polymyxin B, and tigecycline showed the highest susceptibility rates (100%) against the isolates studied, followed by meropenem (57.1%), imipenem (14.3%), and cefepime (14.3%). In contrast, 100% of the isolates were resistant to ertapenem, ceftriaxone, and aztreonam. Resistance to ceftazidime, cefoxitin, and ciprofloxacin was observed in six isolates (87.5%). All isolates showed positive MHT results and were found to carry the blaKPC-2 gene. Two PFGE patterns, A and B, were found among the seven KPC-KPN; the genotype A was the most frequent (n = 5; 71.3%). Besides the presence of blaKPC-2, genotype A (ST 437) isolates also carried blaTEM-1, blaOXA-1, blaCTX-M-15, and blaSHV-11, while genotype B (ST 70) carried only blaSHV-27. Using the dendrogram, a similarity of nearly 79.3% was detected between genotype A and B. The similarity of the KPC-producing isolates evaluated in this study compared to those previously isolated in Recife3 was 74.5%.
Clinical features of the patients who had K. pneumoniae KPC-2-producing isolates at Hospital University Lauro Wanderley, João Pessoa, Paraíba, Brazil.
| Patient | Isolate | PFGE pattern | ST | Gender | Age (y) | Underlyning disease | Primary infection | Treatment | Antimicrobial resistant | MIC carbapenems (CLSI 2010)* | Bacteria isolation (day/mo/yr) | Beta-lactamases | ICU admission (day/mo/yr) | Death (day/mo/yr) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMI | MEM | ||||||||||||||
| 1 | 335 | A1 | 437 | F | 52 | Bowel cancer | Bloodstream | CS, MEM, AMG | ERT, CAZ, CRO, CFO, CIP, ATM | 1 (S) | 1 (S) | 20/02/2010 | KPC-2; TEM-1; OXA-1; CTX-M-15; SHV-11 | 19/02/2010 | 22/02/2010 |
| 2 | 336 | A1 | 437 | F | 71 | Diabetes | Bloodstream | CAZ, CLI, CIP, MEM | ERT, CAZ, CRO, CFO, CIP, ATM | 2 (I) | 2 (I) | 09/09/2009 | KPC-2; TEM-1; OXA-1; CTX-M-15; SHV-11 | 03/09/2009 | 09/09/2009 |
| 339 | A1 | 437 | F | 71 | Diabetes | Urinary tract | ERT, IMI, CAZ, CRO, CFO CIP, ATM | 16 (R) | 0,5 (S) | ||||||
| 3 | 337 | B1 | 70 | F | 29 | HIV | Bloodstream | CS, MEM, AMG, FUN | ERT, CRO, CIP, ATM | 2 (I) | 1 (S) | 13/02/2009 | KPC-2; SHV-27 | 21/02/2009 | 16/03/2009 |
| 338 | B1 | 70 | F | 29 | HIV | Pneumonia | ERT, CAZ, CRO, CFO, ATM, MEM, IMI | 8 (R) | 64 (R) | 20/02/2009 | |||||
| 4 | 340 | A1 | 437 | M | 65 | Encephalopaty | Bloodstream | CS, MEM, AMG, VAN | ERT, CAZ, IMI, CRO, CFO, CIP, ATM | 4 (R) | 2 (I) | 08/08/2009 | KPC-2; TEM-1; OXA-1; CTX-M-15; SHV-11 | 18/08/2009 | 25/10/2009 |
| 5 | 341 | A1 | 437 | F | 52 | Post gastrectomy | Wound | MEM | ERT, CAZ, IMI, CRO, CFO, CIP, ATM | 4 (R) | 1 (S) | 17/08/2009 | KPC-2; TEM-1; OXA-1; CTX-M-15; SHV-11 | 12/08/2009 | 18/08/2009 |
AMG, aminoglycosides; ATM, aztreonam; CAZ, ceftazidime; CFO, cefoxitin; CIP, ciprofloxacin; CLI, clindamycin; CRO, ceftriaxone; CS, 3th and 4th cephalosporins; ERT, ertapenem; F, female; FUN, antifungal; HIV, human immunodeficiency virus infection; I, susceptibility reduced; ICU, intensive care unit; IMI, imipenem; M, male; MEM, meropenem; R; resistant; S, susceptible; ST, sequence type; VAN, vancomycin.
For the first time, K. pneumoniae isolates resistant to ertapenem were detected in the Hospital Lauro Wanderley, which were further confirmed as KPC producers. Among the isolates studied, one (335) showed susceptibility to meropenem and imipenem, whereas three isolates (337, 339, and 341) were only susceptible to meropenem, according to the CLSI breakpoints.10 The discrepancy observed in the susceptible categories to carbapenems may be attributed to production of carbapenemases like KPC. The co-production of KPC with other beta-lactamases, such as TEM-1, OXA-1, SHV-11, SHV-27 and CTX-M-15, was observed among KPC-KPN isolates. These findings are in agreement with previous Brazilian reports which showed that KPC-KPN isolates also possessed the blaCTX-M, blaTEM, and blaSHV genes.3,5,15 Although CTX-M-2 is a widespread variant in South America, the isolates evaluated carried blaCTX-M-15, a frequently found worldwide CTX-M-type. Additionally, according to the authors’ knowledge, this is the first description of co-production of KPC-2 and OXA-1 in Brazilian isolates. ST 437, the most prevalent among the KPC-KPN evaluated, is a ST related to the clonal complex 258, which is widely disseminated among KPC-KPN in Brazil, and associated with the dissemination of KPC worldwide.
Since the therapeutic options are limited and the appropriate empirical antimicrobial treatment is of crucial importance to patient outcome,16 we suggest that the clinical laboratory perform accurate susceptibility testing for KPC producers, including the determination of MICs for tigecycline, aminoglycosides, polymyxins, and carbapenems, since KPC-KPN isolates with low carbapenems MICs and/or isolates with discrepancy for susceptible category among carbapenems tested were observed. In addition, regional surveillance studies that monitor the dissemination of ESBL and carbapenemase enzymes are crucial, since most of these genes are located on mobile genetic elements, which are easily transferred to other bacteria species. Because four of the five isolates were clonally related, suggesting a patient-to-patient transmission, implementation of infection control measures is necessary to restrain the dissemination of resistant genes.
Conflict of interestAll authors declare to have no conflict of interest.
The authors would like to thank Vinícius G. S. Oliveira and Adriana G. Nicoletti for performing the PFGE and MLST assays, respectively, and Ana Carolina R. Silva for her technical assistance with the PCR and sequencing.
Part of this study was presented at the II International Symposium of Clinical Microbiology, in Florianópolis, Brazil, September 29 to October 2, 2010.