Bloodstream infections (BSIs) are among the most concerning bacterial infections. They are one of the leading causes of morbidity and mortality, and occur in 30–70% of critical care patients. The prompt identification of the causative microorganism can help choosing the appropriate antimicrobial therapy that will lead to better clinical outcomes. Blood culture is one of the most relevant tests for microbiological diagnosis of bacterial infections. The introduction of the MALDI-TOF microbiological diagnosis significantly decreased the time of identifying microorganisms. However, it depends on the growth on solid culture medium. In this study, 538 bottles of positive blood cultures were evaluated to test the accuracy of an in house modified protocol. The study sample consisted of 198 Gram-negative and 350 Gram-positive bacteria. In all, 460 (83.94%) species were identified based on the direct plate findings. The protocol allowed the identification of 185/198 (93.43%) of the Gram-negative bacteria, including aerobes, anaerobes, and non-fermenters, and 275/350 (78.85%) of the Gram-positive bacteria. The proposed method has the potential to provide accurate results in comparison to the traditional method with the potential to reduce the turnaround time for the results and optimize antimicrobial therapy in BSI.
Bloodstream infections (BSIs) are one of the leading causes of admittance to intensive care units,1 afflicting up to 2% of inpatients.2 As many as 19 million cases are reported yearly worldwide,3 which makes BSIs one of the leading causes of morbidity and mortality. It occurs in 30–70% among intensive care patients, especially when an antimicrobial effective therapy is not readily instituted.4–6 Sepsis is caused by a variety of microorganisms, and timely diagnosis is of extreme importance due to the severity of the patient's condition, which demands prompt and appropriate antimicrobial therapy.7 In most cases, immediate empirical treatment is initiated, which must be tailored to the microorganism recovered from blood cultures and on antibiotic susceptibility testing. Although the introduction of automated systems in the routine of microbiology laboratories has led to a decrease in turnaround time, the delay in the identification of microorganisms implicated in BSIs is still considerable, given the severity of the patient's status. The microbiological diagnosis of bacteremia consists of performing blood test positivation after the sample is Gram stained. It is then followed by subcultivation of a solid medium and testing of antibiotic susceptibility that takes 12–48h on average depending on the microorganism.8 In view of the delay observed between sample collection and availability of blood culture results, new methods based on molecular biology have been developed and incorporated into laboratory routines, but they are considered limited and expensive.9 ‘Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS)’ introduced in microbiology since 1995, is regarded as a revolutionary method in clinical microbiology, because it identifies microorganisms quickly and accurately, including bacteria and fungi.8,10 The main use of MALDI-TOF is in the identification of microorganisms growing in culture medium, which takes 18–24h. This is a classical method diffused in most microbiology centers. In order to accelerate the identification of microorganisms, various research protocols for yeast fungi and bacteria have been developed directly from positive blood culture vials within two to five hours, depending on the technique used. This study sought to improve the in house protocol through the experience of other studies, developing a method capable of identifying bacteria and yeast fungi from positive blood cultures using mass spectrometry (MALDI TOF), obtaining results in less than two hours. Thus, this protocol significantly reduces the time required for identification of the etiologic agent, reduces the costs of the test, and optimizes the early use of appropriate antimicrobials.5–8
A total of 538 positive bottles Monitored by Automating BacT/ALERT® 3D (bioMérieux, Marcy l’Etoile, France) were analyzed at São Rafael Hospital's Microbiology Laboratory in Salvador, Bahia, Brazil, between May and November 2014. Identifying microorganisms directly from a positive blood culture bottle was compared to the identification after growth in culture medium plates using the MALDI-TOF method. The technique consisted of adding 1mL of the positive blood sample to Eppendorf tube with 200μL of 2% saponin, followed by vortexing for a period of 30s and incubation for 5min at room temperature. The sample was then centrifuged at 14,500×g (1min) and the supernatant was discarded. The pellet was homogenized in 1mL of sterile distilled water, vortexed to complete dissolution, and centrifuged at 14,500×g for 1min. Washing of the pellet in distilled water was repeated twice in accordance with the adapted protocol.11 Subsequently, the supernatant was removed and spots were made from the homogenized pellet. After air-drying at room temperature, each spot was overlaid with 0.5μL of formic acid (bioMérieux) and 1μL of the matrix solution (α-cyano-4-hydroxycinnamic acid) (bioMérieux) was deposited onto the spots. After the spots were air-dried, the target slide was analyzed by the VITEK® MS system.
Finally, the results were compared with those obtained from the growth of the microorganism in the culture medium identified by MALDI-TOF (VITEK® MS, bioMérieux). The results using the protocol of identification directly from the positive blood culture bottles were compared to those obtained with the identification of the same microorganism isolated from a solid medium after subcultivation of the positive bottle using MALDI-TOF. Quality control was performed across readings using the reference E. coli ATCC 8739 strain.
The agreement rate was calculated based on the proportion of identical results obtained using the protocol and direct identification of cultures for each specimen. Partial agreement was considered when the protocol identified at least one of the microorganisms detected by the direct method of the culture plate. Sensitivity and specificity were calculated by considering the culture as gold-standard for true positive and negative findings for each species. The inter-method agreement beyond chance was assessed by calculating Fleiss's Kappa Coefficient. The proportion of correct identification per species and group (Gram-positive and Gram-negative) was also determined. The analyses were performed using the R programming language and environment.
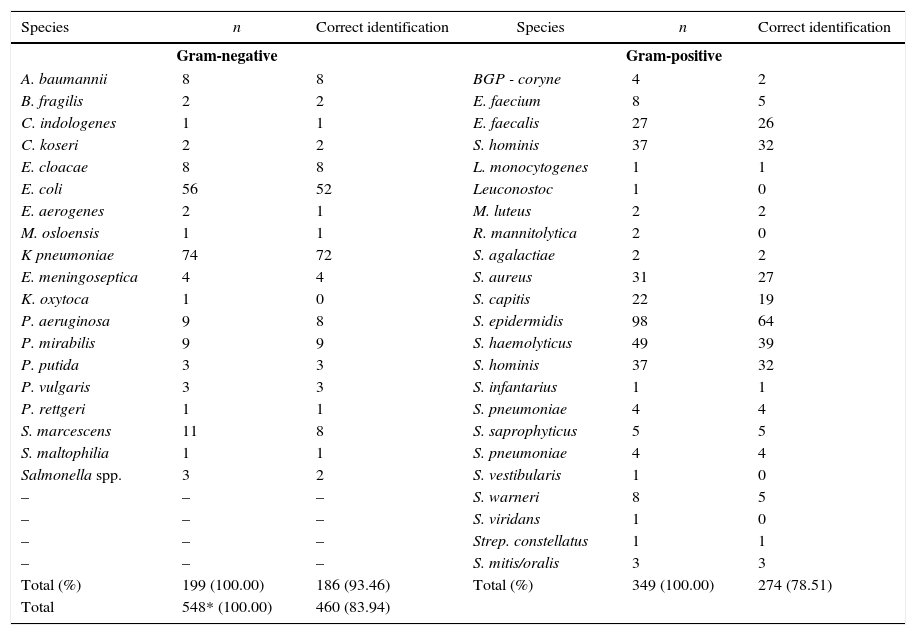
During the study period, 538 positive blood cultures were analyzed using both microorganism identification methods. The sample was predominantly (90.9%) monomicrobial, with a smaller proportion (2.1%) of polymicrobial cultures. Only 7% of the bottles were negative and showed no growth on the culture medium. These were considered false positive results. Most (63.7%) of the 460 microorganisms identified (83.9% of the total) were Gram-positive with all the samples with a score value>90%. Out of 88 incongruous samples, 60 (68.2%) were not detected and 28 (31.8%) were misidentified. The highest percentage of correctly identified microorganisms, in terms of genera and species levels, was found among the Gram-negative bacteria, with 186/199 (93.5%) versus 274/349 (78.5%) of Gram-positive bacteria (Table 1). A higher sensitivity (84.7%; 95% CI: 81.6%–87.8%) than specificity (77.5%; 95% CI: 64.6%–90.4%) was observed, although the confidence interval of the latter was larger due to the small proportion of negative cultures in the study sample. Agreement analysis showed an overall agreement of 81.4% (95% CI: 78.1%–84.7%), with a kappa of 0.798 (95% CI: 0.762–0.834). Agreement increased to 84.2% (95% CI: 81.1%–87.3%). Partial identifications were observed in 11 samples. In nine of them, the culture showed one additional species whereas in two samples the MALDI-TOF method identified one additional species.
Proportion of correct identification per species and group.
| Species | n | Correct identification | Species | n | Correct identification |
|---|---|---|---|---|---|
| Gram-negative | Gram-positive | ||||
| A. baumannii | 8 | 8 | BGP - coryne | 4 | 2 |
| B. fragilis | 2 | 2 | E. faecium | 8 | 5 |
| C. indologenes | 1 | 1 | E. faecalis | 27 | 26 |
| C. koseri | 2 | 2 | S. hominis | 37 | 32 |
| E. cloacae | 8 | 8 | L. monocytogenes | 1 | 1 |
| E. coli | 56 | 52 | Leuconostoc | 1 | 0 |
| E. aerogenes | 2 | 1 | M. luteus | 2 | 2 |
| M. osloensis | 1 | 1 | R. mannitolytica | 2 | 0 |
| K pneumoniae | 74 | 72 | S. agalactiae | 2 | 2 |
| E. meningoseptica | 4 | 4 | S. aureus | 31 | 27 |
| K. oxytoca | 1 | 0 | S. capitis | 22 | 19 |
| P. aeruginosa | 9 | 8 | S. epidermidis | 98 | 64 |
| P. mirabilis | 9 | 9 | S. haemolyticus | 49 | 39 |
| P. putida | 3 | 3 | S. hominis | 37 | 32 |
| P. vulgaris | 3 | 3 | S. infantarius | 1 | 1 |
| P. rettgeri | 1 | 1 | S. pneumoniae | 4 | 4 |
| S. marcescens | 11 | 8 | S. saprophyticus | 5 | 5 |
| S. maltophilia | 1 | 1 | S. pneumoniae | 4 | 4 |
| Salmonella spp. | 3 | 2 | S. vestibularis | 1 | 0 |
| – | – | – | S. warneri | 8 | 5 |
| – | – | – | S. viridans | 1 | 0 |
| – | – | – | Strep. constellatus | 1 | 1 |
| – | – | – | S. mitis/oralis | 3 | 3 |
| Total (%) | 199 (100.00) | 186 (93.46) | Total (%) | 349 (100.00) | 274 (78.51) |
| Total | 548* (100.00) | 460 (83.94) | |||
Total number of bacteria (548) is bigger than the total number of sample due to polymicrobial cultures.
This study analyzed positive blood cultures of patients with bacteremia including Gram-positive and Gram-negative bacteria. It compared direct microorganism identification from positive bottles to growth on solid medium using MALDI-TOF MS. The latter relies on the detection of ribosomal proteins and is considered a fast and accurate method for microbiological diagnosis. It is described in a great number of studies as an important tool to aid physicians in choosing the appropriate antimicrobial therapy, thereby reducing potential adverse events in patients with BSI.1 Our protocol showed agreement in the identification of the microorganism grown on a solid medium. In relation to established protocols, similar or superior agreement was obtained, regardless of the processing periods such as the one in a study conducted by Martiny et al., 2012.11 Using an in-house method, they found 73.7% of correct identifications overall, with 86.4% correctly identified Gram-negative bacteria and 73.7% Gram-positive genera and species. Moussaouil et al., 2010, showed correct identification of 89.7% of the bottles12; Drancourt, 2010, found 94.0% for Gram-negative and 67.0% for Gram-positive bacteria13; and La Scola & Raoult, 2009, obtained 88.0% correctly identified Gram-negatives and 67.0% Gram-positives.8
The present study yielded 84% of correct identifications at the species level, with 93.5% being Gram-negative and 78.5% Gram-positive bacteria. Sensitivity (77.5%) and specificity (84.7%) values were consistent with those widely found in medical literature, and can assist microbiologists in their decision-making regarding the identification method to use. In spite of the controversies around Kappa measurement of agreement classifications, the study showed a kappa of 0.798 – which reflects substantial agreement according to most of the literature.14,15
Our study showed reliable and accurate results when using MALDI-TOF compared to conventional methods. Our in house protocol is considered to be an efficient and successful method for routine identification of bacterial isolates in clinical microbiology; unlike other standardized protocols, which are described as lengthy and as difficult to be deployed in routine microbiology.
Conflicts of interestThe authors declare no conflicts of interest.