Mitogen-activated protein kinase (MAPK) signaling pathway plays an important role in response to viral infection. The aim of this study was to explore the function and mechanism of MAPK signaling pathway in enterovirus 71 (EV71) infection of human rhabdomyosarcoma (RD) cells.
MethodsApoptosis of RD cells was observed using annexin V-FITC/PI binding assay under a fluorescence microscope. Cellular RNA was extracted and transcribed to cDNA. The expressions of 56 genes of MAPK signaling pathway in EV71-infected RD cells at 8h and 20h after infection were analyzed by PCR array. The levels of IL-2, IL-4, IL-10, and TNF-α in the supernatant of RD cells infected with EV71 at different time points were measured by ELISA.
ResultsThe viability of RD cells decreased obviously within 48h after EV71 infection. Compared with the control group, EV71 infection resulted in the significantly enhanced releases of IL-2, IL-4, IL-10 and TNF-α from infected RD cells (p<0.05). At 8h after infection, the expressions of c-Jun, c-Fos, IFN-β, MEKK1, MLK3 and NIK genes in EV71-infected RD cells were up-regulated by 2.08–6.12-fold, whereas other 19 genes (e.g. AKT1, AKT2, E2F1, IKK and NF-κB1) exhibited down-regulation. However, at 20h after infection, those MAPK signaling molecules including MEKK1, ASK1, MLK2, MLK3, NIK, MEK1, MEK2, MEK4, MEK7, ERK1, JNK1 and JNK2 were up-regulated. In addition, the expressions of AKT2, ELK1, c-Jun, c-Fos, NF-κB p65, PI3K and STAT1 were also increased.
ConclusionEV71 infection induces the differential gene expressions of MAPK signaling pathway such as ERK, JNK and PI3K/AKT in RD cells, which may be associated with the secretions of inflammatory cytokines and host cell apoptosis.
Human enterovirus belongs to the RNA virus family Picornaviridae which includes polioviruses, type A coxsackieviruses, type B coxsackieviruses, echoviruses, and enterovirus types 68–71. Among these viruses, human EV71 and coxsackievirus A16 (CVA16) are major causative agents of hand, foot, and mouth diseases (HFMD).1 In recent years, the mortality rate of EV71 infection was significantly elevated, and there are neither approved therapies nor vaccines for EV71.2,3 Although the underlying mechanisms remain obscure, host innate immunity is thought to play an important role in virus infection.4,5 MAPKs are key signaling molecules in innate immunity, which are a family of serine/threonine protein kinases widely conserved in eukaryotes and involved in many cellular functions such as inflammation, cell proliferation, differentiation, movement, and death.6–10 To date, seven distinct MAPK groups have been characterized in mammalian cells: extracellular regulated kinases (ERK1/2), c-Jun NH2 terminal kinases (JNK1/2/3), p38 MAPK (p38 α/β/γ/δ), ERK3/4, ERK5, ERK7/8, and Nemo-like kinase (NLK).11,12 Among these, the most extensively studied groups are ERK1/2, JNKs and p38 MAPK. Virus-infected cells induce acute reactions that not only activate MAPK signaling cascades to defend virus infection but also can be used by viruses to support viral replication.13 Previous studies have shown that influenza virus, herpes simplex virus type 2 and human immunodeficiency virus type 1 (HIV-1) can activate MAPKs including ERK1/2 (p42/p44 MAPK), JNK, or p38 MAPK, which further increase the expressions of different cytokines or affect the growth of viruses.14–16 In addition, many genes regulated by MAPKs are dependent on NF-κB, c-Fos and c-Jun for transcription, thus activating the gene expressions of pro-inflammatory or antiviral cytokines to release IL-6, IFN-β and TNF-α.17,18 In this study, we employed PCR array to explore the differential gene expressions of MAPK signaling pathway proteins in EV71-infected RD cells, and demonstrated that the releases of IL-2, IL-4, IL-10 and TNF-α may be associated with the activation of ERK, JNK and PI3K/AKT.
Materials and methodsVirus preparation and plaque assayRD cells were purchased from CBTCCCAS (Chinese Academy of Sciences Cell Bank of Type Culture Collection) and cultured in high glucose DMEM (Thermo Scientific HyClone, UT, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C in a humidified incubator under 5% CO2 atmosphere. EV71 strain CCTCC/GDV083 (ATCC VR-784) (China Center for Type Culture Collection, CCTCC) was propagated using RD cells. Once 90% of the cells showed cytopathic effect (CPE), the viral supernatants were gathered and centrifuged at 4000rpm for 5min. The clear supernatant was then transferred to a new tube, and the debris was frozen and thawed three times before centrifugation. Finally, the supernatants were pooled and stored at −70°C. RD cells (4×105cells per well) were seeded into a six-well culture plate overnight and then infected with a 10-fold serially diluted virus suspension. One hour after infection, the cells were washed once with PBS and overlaid with 0.3% agarose in DMEM containing 2% FBS. After 96h incubation, the cells were fixed with 10% formaldehyde and then stained with 1% crystal violet solution. The virus titers were determined by plaque assays using RD cells in triplicate.
EV71 infection and viability of RD cellsOne batch of uninfected RD cells in 25cm2 culture flask were used as the control, while two other batches of RD cells were infected with UV-inactivated EV71 strain and alive EV71 strain at a multiplicity of infection (MOI) of 5 in 4mL of virus inoculum diluted with maintenance medium. Approximately 1×106cells were incubated with EV71 at an MOI of 5 or as indicated and allowed to absorb for 2h at 37°C. Unbound viruses were removed by washing the cells with medium, and 15mL of maintenance medium was added. Infected cells and culture supernatants were collected at different time intervals. The viability of RD cells was assayed using trypan blue exclusion method.
Evaluation of apoptosis in RD cells by annexin V-FITC/PI binding assayRD cells were seeded at a density of 1×105cells onto pre-treated coverslips in a 6-well plate. After incubation at 37°C overnight, the cells were inoculated at an MOI of 5 with EV71 strain and UV-inactivated EV71 strain for 2h. After absorption, the inoculum was removed, and replaced with 3mL of maintenance medium. The cells were subjected to incubation for another 8h, and then fixed with 3.7% paraformaldehyde for 30min at room temperature. The coverslips were stained by annexin V-FITC/PI (Major BioTech, Shanghai, China) and observed under a fluorescence microscope.
Detection of cytokines by ELISACulture supernatants were collected at 0h, 20h and 32h after EV71 infection (MOI=5), including uninfected control and mock-infected control, and then stored at −70°C until use. ELISA kits for IL-2, IL-4, IL-10 and TNF-α were purchased from Westang Biotechnology (Shanghai, China) and the ELISA assay was performed according to the manufacturer's protocol. Spectrometric absorbance at wavelength of 450nm was measured on an enzyme immunoassay analyzer (Model 680, Bio-Rad, USA). All assays were performed in triplicate.
RNA isolation and PCR arrayAfter incubation at 37°C for 8h and 20h, both uninfected and infected RD cells (MOI=5) were harvested for the extraction of total RNA. RNA was extracted from RD cells with SV total RNA isolation system (Promega, Madison, WI, USA) according to the manufacturer's instructions. Reverse transcription was performed using RT-PCR Kit (CT biosciences; catalog# CTB101, China) and then run on the ABI 9700 thermocycler (ABI, Foster City, CA). A total of 56 kinds of primers including p38 MAPK, JNK, ERK and NF-κB were designed according to gene sequences published in GenBank. Gene-specific primers for B2M, ACTB, GAPDH, RPL27, HPRT1 and OAZ1, as well as a PCR positive control and a genomic DNA control were fixed in 96-well PCR plate. PCR were performed with customized PCR arrays containing pre-dispensed primer assays (CT biosciences, China) on the LightCycler 480 (Roche Diagnostics, Mannheim, Germany) using SYBR MasterMix (CT biosciences; catalog# CTB103, China). Each PCR contained cDNA synthesized from 10ng of total RNA. The PCR amplification parameters were 95°C for 5min, followed by 40 cycles of 95°C for 15s and 60°C for 15s and 72°C for 20s. Two PCR arrays for EV71-infected RD cells and uninfected RD cell control were used for relative quantification of gene expression. This experiment was repeated three times. Fold change was calculated using the formula of 2-ΔΔCt.
Statistical analysisAll data were presented as the mean±SE. Statistical analyses were preformed using GraphPad Prism software (San Diego, CA). The three groups were compared using two-way ANOVA. A p-value less than 0.05 was considered statistically significant.
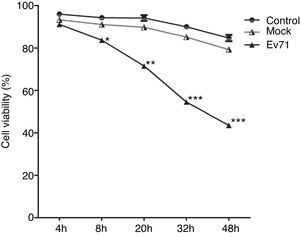
ResultsGrowth characteristics of EV71-infected RD cellsTwo batches of RD cells with three experimental models in each group including uninfected RD cells (Control), UV-inactivated EV71-infected RD cells (Mock), and EV71-infected RD cells (EV71) were analyzed at 4h, 8h, 20h, 32h and 48h after infection. In uninfected control and mock-infected control, the growth of the cells did not exhibit an obvious difference at 4h and 8h. However, the viable EV71-infected cells exhibited a significant decrease in growth at 20h, 32h and 48h (Fig. 1). Visible CPE was also observed in the infected cell culture (Fig. 2).
EV71 infection induced RD cell apoptosis. The viability of uninfected RD cells (Control), UV-inactivated EV71-infected RD cells (Mock), and EV71-infected RD cells (EV71) were determined at designated time points after EV71 infection by trypan blue exclusion technique. The data were expressed as mean±SE from three independent experiments and analyzed by two-way ANOVA with Bonferroni post hoc tests. *p<0.05, **p<0.01, and ***p<0.001.
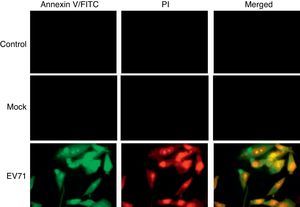
The phosphatidylserine (PS) was localized mainly on the membrane surface of cytoplasmic cells. However, PS could be translocated to the outer leaflet of the membrane during apoptosis. Staining cells with annexin V-FITC allowed for the detection of PS exposure as an early indicator of apoptosis. When RD cells were infected with EV71 at 8h, the cells were stained with annexin V-FITC/PI and visualized under a fluorescence microscope (Fig. 3).
EV71-induced apoptosis in RD cells. Control: uninfected RD cells; Mock: UV-inactivated EV71-infected RD cells. EV71: EV71-infected RD cells. Early membrane revealed an obvious change associated with apoptosis in EV71-infected RD cells. The exposure of phosphatidylserine (PS) was analyzed using FITC-labeled annexin V. Cell death was assessed with propidium iodide (PI). EV71-infected RD cells were harvested and stained with annexin V-FITC/PI at 8h after infection, and then examined under a fluorescence microscope (1000×).
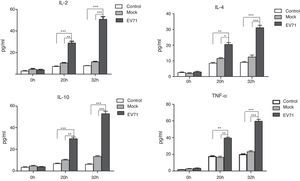
Culture supernatants were harvested from the control, mock-infected or EV71-infected RD cells at 0h, 20h and 32h after infection, and the levels of IL-2, IL-4, IL-10 and TNF-α were measured by ELISA. Once RD cells were infected by EV71, the obviously increased secretion of cytokines such as IL-2, IL-4, IL-10 and TNF-α was observed (Fig. 4). Meanwhile, a significant difference among three groups was also observed.
Cytokine production in EV71-infected RD cells. RD cells were infected with EV71 (MOI=5). At 0h, 20h and 32h, cell culture supernatants were collected and the levels of cytokines were performed by ELISA. The data were expressed as mean±SE from three independent experiments and analyzed by two-way ANOVA with Bonferroni post hoc tests. *p<0.05, **p<0.01, and ***p<0.001.
Eight hours after RD cells were infected with EV71, the expression levels of c-Jun, c-Fos, IFN-β, MEKK1 (MAP3K1), MLK3 (MAP3K11) and NIK (MAP3K14) revealed 2.08–6.12 fold enhancement, while other 19 genes (e.g. AKT1, AKT2, E2F1, IKK and NF-κB1) exhibited down-regulation. However, at 20h after infection, AKT2, ELK1, c-Jun, c-Fos, NF-κB p65, PI3K and STAT1 in EV71-infected RD cells were up-regulated and exhibited 2.19–5.52 fold enhancement in expression. Meanwhile, MAPK signaling pathway molecules including MEKK1, ASK1 (MAP3K5), MLK2 (MAP3K10), MLK3, NIK, MEK1 (MAP2K1), MEK2 (MAP2K2), MEK4 (MAP2K4), MEK7 (MAP2K7), ERK1 (MAPK3), JNK1 (MAPK8) and JNK2 (MAPK9) were significantly up-regulated and revealed 2.00–4.09 fold increases in expression level (Table 1).
Differential gene expressions of MAPK signaling pathway in RD cells in response to EV71 infection at 8h and 20h postinfection.
| Symbols | Description of genes | EV71/control (fold changes) | |
|---|---|---|---|
| 8h | 20h | ||
| Akt1 | V-akt murine thymoma viral oncogene homolog 1 (AKT1) | −9.89 | −1.13 |
| Akt2 | V-akt murine thymoma viral oncogene homolog 2 (AKT2) | −3.36 | 2.66 |
| Akt3 | V-akt murine thymoma viral oncogene homolog 3 (AKT3) | −1.06 | 1.58 |
| c-Fos | c-fos proto-oncogene | 3.34 | 3.23 |
| c-Jun | c-Jun proto-oncogene | 3.19 | 5.52 |
| E2F1 | Transcription factor E2F1 (RBAP1; RBBP3; RBP3) | −3.99 | −1.17 |
| E2F2 | Transcription factor E2F2 | −1.64 | 1.55 |
| ELK1 | E twenty-six (ETS)-like transcription factor 1 | 1.19 | 2.19 |
| EGFR | Epidermal growth factor receptor | −4.78 | 1.38 |
| IFN-β | Interferon beta | 5.22 | 1.47 |
| IGF1 R | Insulin-like growth factor 1 (IGF-1), receptor (CD221; IGFR; JTK13) | −3.50 | 1.27 |
| IKBKB | Inhibitor of nuclear factor kappa-B kinase subunit beta (IKK-β; IKKB) | −3.52 | 1.33 |
| IKBKG | Inhibitor of nuclear factor kappa-B kinase subunit gamma (FIP3; IKK-γ; NEMO) | −5.76 | 1.40 |
| IL-1α | Interleukin-1 alpha | −2.60 | 1.64 |
| IL-1β | In Interleukin-1 beta | −2.20 | 1.30 |
| IL-2 | Interleukin 2 | 1.24 | 12.15 |
| IL-4 | Interleukin 4 | −3.72 | 2.39 |
| IL-6R | Interleukin 6 receptor (CD126) | −5.80 | 1.92 |
| IL-7 | Interleukin 7 | −1.65 | 1.26 |
| IL-10 | Interleukin 10 | 1.10 | 12.15 |
| IRAK1 | Interleukin-1 receptor-associated kinase 1 | −3.70 | 3.39 |
| JAK2 | Janus kinase 2 (JTK10; THCYT3) | 1.06 | −1.21 |
| MADD | MAP-kinase activating death domain | 1.57 | 1.82 |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 (MEKK 1; MAPKKK 1; SRXY6) | 2.08 | 2.25 |
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 (ASK1; MAPKKK5; MEKK5) | 1.30 | 2.00 |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 (MEKK7; MAPKKK 7;TAK1) | −1.13 | 1.14 |
| MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 (MLK2) | 1.48 | 4.07 |
| MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 (MEKK11; MLK3; PTK1) | 2.61 | 3.85 |
| MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 (MEKK14, NIK) | 6.12 | 4.09 |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 (MAPKK1; MEK1) | 1.30 | 2.46 |
| MAP2K2 | Mitogen-activated protein kinase kinase 2 (MAPKK2; MEK2) | −1.31 | 2.18 |
| MAP2K3 | Mitogen-activated protein kinase kinase 3 (MAPKK3; MEK3) | −2.29 | −1.10 |
| MAP2K4 | Mitogen-activated protein kinase kinase 4 (JNKK1; MAPKK4; MEK4) | −1.06 | 2.54 |
| MAP2K6 | Mitogen-activated protein kinase kinase 6 (MAPKK6; MEK6) | 1.58 | −1.23 |
| MAP2K7 | Mitogen-activated protein kinase kinase 7 (MAPKK7; MEK7) | −1.09 | 3.05 |
| MAPK1 | Mitogen-activated protein kinase 1 (ERK2; MAPK2; P42-MAPK) | 1.20 | 1.00 |
| MAPK3 | Mitogen-activated protein kinase 3 (ERK1; P44-MAPK) | 1.67 | 3.95 |
| MAPK8 | Mitogen-activated protein kinase 8 (JNK; JNK1; SAPK1) | 1.73 | 2.53 |
| MAPK9 | Mitogen-activated protein kinase 9 (JNK2; JNK2A; JNK2B; SAPK) | 1.02 | 2.25 |
| MAPK10 | Mitogen-activated protein kinase 10 (JNK3; SAPK1b) | 1.56 | 1.30 |
| MAPK11 | Mitogen-activated protein kinase 11 (p38-β MAPK) | 1.80 | 1.01 |
| MAPK12 | Mitogen-activated protein kinase 12 (p38-γ MAPK, ERK6) | 1.13 | −1.00 |
| MAPK13 | Mitogen-activated protein kinase 13 (p38-δ MAPK, SAPK4) | 1.52 | −1.34 |
| MAPK14 | Mitogen-activated protein kinase 14 (p38-α MAPK) | 1.15 | −1.19 |
| NF-κB1 | Nuclear factor NF-kappa-B p105 subunit (NF-kB1; NF-κB-p105; NF-κB-p50) | −4.81 | 1.18 |
| NF-κB3 | Nuclear factor NF-kappa-B p65 (RELA; NF-κB p65) | −1.35 | 2.63 |
| PI3Kα | Phosphoinositide-3-kinase, catalytic, alpha polypeptide (PI3K; p110-alpha) | −1.26 | 1.04 |
| PI3Kγ | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3CG; PI3K; PI3Kgamma) | −1.40 | 5.18 |
| PIK3R2 | Phosphatidylinositol 3-kinase regulatory subunit beta (P85B; p85; p85-BETA) | −2.12 | −1.20 |
| PPP3R1 | Protein phosphatase 2B regulatory subunit 1 (CALNB1; CNB1) | −1.52 | 1.94 |
| RIPK1 | Receptor-interacting serine/threonine-protein kinase 1 (RIP; RIP1) | −2.44 | 1.24 |
| STAT1 | Signal Transducers and Activators of Transcription (CANDF7; ISGF-3; STAT91) | −6.18 | 4.04 |
| STAT5A | Signal transducer and activator of transcription 5A | −1.14 | −1.15 |
| STAT5B | Signal transducer and activator of transcription 5B | −3.17 | 1.08 |
| TGF-β1 | Transforming growth factor beta 1 (DPD1; LAP; TGFbeta) | −2.23 | −1.15 |
| TNF-α | Tumor necrosis factors | −1.97 | 2.19 |
Negative values indicate down-regulation of genes.
MAPKs are a heterogeneous group of enzymes responsible for phosphorylating serine and threonine in many proteins.19,20 In mammals, three major MAPK signaling pathways have been identified as ERK, JNK and p38 MAPK.21,22 MAPKs are phosphorylated and activated by MAPK-kinases (MAPKKs or MAP2Ks), which in turn are phosphorylated and activated by MAPKK-kinases (MAPKKK or MAP3Ks).23 As previously reported, many viruses including HIV, hepatitis C virus (HCV) and Kaposi's sarcoma-associated herpesvirus (KSHV) can target MAPK pathways as a means to manipulate cellular function and to control viral infection and replication.24–26 In this study, MAPK signaling pathway molecules, including MEKK1, ASK1, MLK, NIK, MEK1/2, MEK4/7, ERK and JNK were significantly up-regulated in EV71-infected RD cells, which stimulated and induced the immune response of RD cells.
ERK was the first MAPK to be identified as the best studied mammalian MAPK pathway.27 ERK1 and ERK2 are two isoforms with 83% amino acid homology. Both isoforms are expressed in various tissues, and can be activated by a large number of extracellular and intracellular stimuli.28 Activated Raf (or other MAP3K) binds and phosphorylates downstream kinase MEK1 and MEK2 with dual specificity, which in turn phosphorylate ERK1/2.29 In this study, the expressions of MEK1, MEK2 and ERK1 genes were significantly up-regulated and revealed 2.46-, 2.18- and 3.95-fold enhancement after EV71 infection, which may be associated with the activation of downstream molecules such as NF-κB p65, ELK-1, c-Fos, c-Jun and STAT1. Whereas, STAT1 is a member of signal transducers and activators in transcription family, and involved in the up-regulation of the genes through type I, type II or type III interferon.30,31 Both NF-κB and STAT1 are rapidly activated in response to various stimuli including viral infection and cytokines, thus controlling the expression of anti-apoptotic, pro-proliferative and immune response genes.32 PCR array showed that the expressions of IL-2, IL-4, IL-10 and TNF-α exhibited the enhancement by 12.15-, 2.39-, 12.15- and 2.19-fold at 20h after infection. Compared with control and inactive EV71-infected Mock, the levels of IL-2, IL-4, IL-10 and TNF-α at 20h and 32h after infection showed significant increases in EV71-infected RD cells (p<0.05, p<0.01, or p<0.001). The results indicated that EV71 infection could increase the releases of IL-2, IL-4, IL-10 and TNF-α in RD cells, as well as mediate inflammation and apoptosis.
Mammalian JNKs are encoded by three distinct genes (JNK1, JNK2 and JNK3). JNK1 and JNK2 are ubiquitously expressed, while JNK3 is restricted to brain, heart and testis tissues.33 JNK pathway is activated by a large number of external stimuli, and the initial signaling pathway starts with the activation of several MAPKKKs including TAK1, MEKK1/4, MLK-2 and -3, and ASK1, which phosphorylate MEK4 or MEK7 and induce the activation of JNK. As mentioned previously, several transcription factors are downstream proteins especially c-Fos and c-Jun activated by JNK.23 Once activated by phosphorylation, these transcription factors can induce the secretions of proinflammatory cytokines.34 In this study, the up-regulation of MEKK1, ASK1, MLK2, MLK3, NIK, MEK4 and MEK7 was observed, which may be related to the activation of JNK signaling pathway. Subsequently, transcription factors of NF-κB p65, ELK-1, c-Fos, c-Jun and STAT1 are activated, which plays a key role in regulating the immune response to viral infection and stimulates the releases of cytokines such as TNF-α, IFN-β, IL-1β, IL-2 and IL-6.35 JNK is also involved in the regulation of apoptosis through the modulation of c-Jun/AP-1 and transforming growth factor-β in MDCK cells.11 Although JNK plays an important role in apoptosis, it also contributes to survival. These opposite effects could be partly due to the duration or magnitude of the pathway activation and the activation of other pro-survival pathways. Prolonged activation of JNK can mediate apoptosis, whereas transient activation can promote cell survival.36
The role of p38 MAPK in immune system has been extensively studied. The p38 MAPK participates in macrophage and neutrophil functional responses including respiratory burst activity, chemotaxis, granular exocytosis, adherence, and apoptosis, as well as mediates T cell differentiation and apoptosis by regulating the production of IFN-γ.37 In addition, p38 MAPKs can be activated by MAPKKs, MKK3 and MKK6. The activation of p38 MAPK controls the expression of RANTES, IL-8 and TNF-α, which appears to control proinflammatory responses after infection.38 In this study, PCR array did not reveal the activation of p38 MAPK signaling pathway genes including p38 MAPK α, β, γ and δ, which may be irrelevant to EV71 infection.
PI3K/AKT signaling pathway plays an important role in the growth and survival of cells. The AKT cascade is activated by receptor tyrosine kinases, cytokine receptors, G-protein coupled receptors, and other stimuli, thus inducing the accumulation of phosphatidylinositol-3,4,5 triphosphates by PI3K.38,39 Three AKT isoforms (AKT1, AKT2 and AKT3) can mediate downstream events through PI3K. As previously reported, EV71 can activate PI3K/AKT to trigger anti-apoptosis pathway at the early phase during infection.40 At 8h after EV71 infection, the expressions of AKT1 and AKT2 were significantly down-regulated and revealed a reduction in expression level by 9.89- and 3.36-fold, while PI3Kγ and AKT2 exhibited an increase by 5.18-fold and 2.66-fold at 20h after EV71 infection, respectively. It is postulated that differential expression of PI3K/AKT in RD cells may be attributed to different conditions such as MOI, infection time, and EV71 strain.
In summary, MAPKs are components of signaling cascades where various extracellular stimuli converge to initiate inflammation, including production of proinflammatory cytokines (e.g. TNF-α, IL-1, IL-2 and IL-6). The releases of most cytokines and chemokines are regulated primarily at the level of transcription via the activation of specific transcription factors controlled by NF-κB and MAPKs. Therefore, the differential gene expressions of ERK, JNK and PI3K/AKT signaling pathway genes may be related to EV71 replication, proinflammatory cytokines and apoptotic responses in RD cells.
Conflict of interestThe authors declare no conflict of interest.
The authors would like to thank Guanghua Luo and Jingting Jiang for their help with flow cytometry and statistical analysis.