The increasing rates of nosocomial infection associated with coagulase–negative staphylococci (CoNS) were the rationale for this study, aiming to categorize oxacillin–resistant CoNS species recovered from blood culture specimens of inpatients at the UNESP Hospital das Clínicas in Botucatu, Brazil, over a 20–year period, and determine their sensitivity to other antimicrobial agents. The mecA gene was detected in 222 (74%) CoNS samples, and the four types of staphylococcal chromosomal cassette mec (SCCmec) were characterized in 19.4%, 3.6%, 54.5%, and 14.4% of specimens, respectively, for types I, II, III, and IV. Minimal inhibitory concentration (MIC) values to inhibit 50% (MIC50) and 90% (MIC90) of specimens were, respectively, 2 and >256μL/mL for oxacillin, 1.5 and 2μL/mL for vancomycin, 0.25 and 0.5μL/mL for linezolid, 0.094 and 0.19μL/mL for daptomycin, 0.19 and 0.5μL/mL for quinupristin/dalfopristin, and 0.125 and 0.38μL/mL for tigecycline. Resistance to oxacillin and tigecycline and intermediate resistance to quinupristin/dalfopristin were observed. Eight (2.7%) of all 300 CoNS specimens studied showed reduced susceptibility to vancomycin. Results from this study show high resistance rates of CoNS to antimicrobial agents, reflecting the necessity of using these drugs judiciously and controlling nosocomial dissemination of these pathogens.
Coagulase-negative staphylococci (CoNS), members of the staphylococci group, are characterized as Gram-positive cocci, presented as single cells or with irregular disposition, and are immobile, non-spore forming, catalase-positive, mostly facultative anaerobes, and lack the enzyme coagulase.1 CoNS are part of the human microbiota, considered opportunistic pathogens, causing infections mostly in premature babies, and immunocompromised and prosthetic patients.2
The rise of antimicrobial resistance in recent years has had a great impact on hospital infections caused by CoNS. Oxacillin is a semisynthetic penicillin used in the susceptibility test for the detection of methicillin resistance and treatment of staphylococcal infections. However, rates from 66% to 95% of oxacillin resistance have been observed in CoNS clinical isolates.2 Oxacillin resistance is often mediated by the mecA gene, which encodes a supplemental penicillin-binding protein (PBP2a) with low-affinity to semisynthetic penicillins.3 The mecA gene is located on a mobile genetic element known as the Staphylococcal Cassette Chromosome mec (SCCmec) which contains the mec complex, composed of the mecA gene and its regulator genes mecI and mecRI, the ccr complex, responsible for integration and excision of the SCCmec, and J region, which is not essential for the SCCmec formation, but may carry non-β–lactam resistance genes.4 To date, 13 SCCmec types have been described, based on the combination of ccr gene complex types and mec gene complex classes. The subtypes were defined by J region polymorphisms in the same combination of mec and ccr complexes.5
The emergence of oxacillin resistant isolates has led to the ultimate use of alternative antimicrobials for treatment of CoNS infections, such as the glycopeptide vancomycin. In the meantime, descriptions of reduced susceptibility and resistance to vancomycin have been reported in recent decades.6 Reduced susceptibility to vancomycin may be related to metabolic modifications such as acceleration in peptidoglycan synthesis, resulting in cell wall thickening. In this process, vancomycin is not able to inhibit the peptidoglycan synthesis, since it is depleted due to the higher availability of D-alanyl-D-alanine sites.7
Given the upsurge in hospital infections caused by CoNS, this study aimed to characterize the oxacillin resistant strains and determine the antimicrobial susceptibility of a 20-year collection of blood culture CoNS isolates from Botucatu Hospital das Clínicas inpatients.
Material and methodsStrainsThree-hundred CoNS strains were isolated from blood cultures from inpatients of the Botucatu Hospital das Clínicas – Paulista State University (UNESP). The isolates were collected from 1990 to 2009 and kept in the Culture Collection Laboratory of the Microbiology and Immunology Department of the Botucatu Biosciences Institute - UNESP. The selection criteria considered a mean prevalence of events of 35%, with a margin of error of 5%, and a 95% confidence interval.
The strains were isolated according to Koneman et al.8 Blood-agar isolates were subjected to Gram stain for observation of colony morphology and the catalase test was performed for confirmation of the genus Staphylococcus. Staphylococcal strains were submitted to the coagulase test for differentiation of the coagulase-negative and coagulase-positive groups. Coagulase-negative isolates were subjected to biochemical tests for the phenotypic identification of species. The genotypic identification was performed using primers drawn over conserved sequences adjacent to 16S and 23S genes, by ITS-PCR (internal transcribed spacer–polymerase chain reaction), described by Couto et al.9 Amplification efficiency was monitored by electrophoresis in 3% metaphor agarose and stained with SYBR Safe. The following lineages of international reference were used: S. auricularis ATCC 33753, S. capitis subsp. capitis ATCC 27843, S. capitis subsp. urealyticus ATCC 49325, S. caprae ATCC 35538, S. cohnii ATCC 49330, S. cohnii subsp. cohnii ATCC 29974, S. epidermidis ATCC 12228, S. epidermidis ATCC 35983, S. hemolyticus ATCC 29970, S. hominis ATCC 27844, S. hominis subsp. novobiosepticus ATCC 700237, S. lentus ATCC 700403, S. lugdunensis ATCC 700328, S. saprophyticus ATCC 15305, S. schleiferi subsp. schleiferi ATCC 43808, S. sciuri subsp. sciuri ATCC 29062, S. simulans ATCC 27851, S. xylosus ATCC 29979, and S. warneri ATCC 10209.
DNA extractionThe Illustra kit (GE Healthcare) was used for DNA extraction. The steps included an initial digestion of staphylococcal cells with lysozyme (10mg/mL) and proteinase K (20mg/mL). Next, 500μL of the extraction buffer were added to the mixture, which was centrifuged at 10,000 x g for 4min. The supernatant was transferred to a column and centrifuged at 5,000g for 1min. The fluid was discarded and 500μL of extraction buffer were added to the column. After the centrifugation and discarding of the collected fluid, 500μL of washing buffer were added to the column, which was submitted to centrifugation at 20,000g for 3min. Next the column was transferred to a 1.5mL tube and elution was performed using 200μL of warmed MilliQ water at 70°C.
The mecA gene detectionPCR was performed for the detection of the mecA gene. Reactions were performed using a protocol described by Murakami et al.10 The amplification efficiency was monitored by electrophoresis in a 2% agarose gel stained with SYBR Safe.
Determination of SCCmecThe SCCmec type was determined on mecA-positive strains. Reactions were performed using a protocol described by Oliveira et al.11 and modified by Machado et al.12
Multiplex PCR was performed in 50μL of reaction volume with 1X enzyme buffer, 1.25 U Taq polymerase DNA, 200 μM dNTP Mix, and the following primers: 10pmol of RIF2 F2 (TTCGAGTTGCTGATGAAGAAGG) and CIF2 R2 (ATTTACCACAAGGACTACCAGC), 6pmol of KDP F1 (AATCATCTGCCATTGGTGATGC) and KDP R1 (CGAATGAAGTGAAAGAAAGTGG), 5pmol of DCS F2 (CATCCTATGATAGCTTGGTC) and DSC R1 (CTAAATCATAGCCATGACCG), 5pmol of RIF4 F3 (GTGATTGTTCGAGATATGTGG), and RIF4 R9 (CGCTTTATCTGTATCTATCGC). For each reaction, 10μL DNA was added. The cycle sequencing reactions were performed at 92°C for 3min, followed by 30 cycles of 92°C for 1min, 56°C for 1min, and 72°C for 1min and 30s. The amplification efficiency was monitored by electrophoresis in a 2% agarose gel stained with SYBR Safe.
Determination of the minimal inhibitory concentration (MIC) by the E-testThe in vitro susceptibility of CoNS strains was tested for the following antimicrobials: Oxacillin, Vancomycin, Daptomycin, Linezolid, Quinupristin/Dalfopristin, and Tigecycline. The MIC of these drugs was determined by the E-test. The criteria used for the susceptibility classification were: Oxacillin <0.5μg/mL (susceptible) for CoNS, except S. lugdunensis (susceptible ≤2μg/mL and resistant ≥4μg/mL); Vancomycin <4μg/mL (susceptible), 8–16μg/mL (intermediate resistant), and >32μg/mL (resistant); Linezolid ≤4μg/mL (susceptible); Daptomycin ≤1μg/mL; Quinupristin/Dalfopristin ≤1μg/mL (susceptible), 2μg/mL (intermediate), and ≥4μg/mL (resistant); Tigecycline ≤0.5μg/mL.13
Screening test for the detection of reduced susceptibility to vancomycinIn order to detect reduced susceptibility to vancomycin, a screening agar test prepared with Brain Heart Infusion (BHI) Agar and 4μg/mL, 6μg/mL, 8μg/mL, and 16μg/mL of vancomycin was used. The reference strain S. aureus ATCC 29213, susceptible to vancomycin, was used as a negative control, and strain E. faecalis ATCC 51299, resistant to vancomycin, as a positive control. Spots of a 2.0 McFarland inoculum were added to the Agar plate and incubated at 35°C for 24h, and the growing of at least one colony was considered as a positive result.
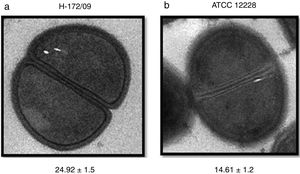
Analysis of the cell wall thicknessThe CoNS strains that presented reduced susceptibility to vancomycin were submitted to the transmission electronic microscopy for cell wall thickness analysis. Strains were cultured in BHI broth and incubated at 37° C for 24h. In a microtube, 1000μL of a CoNS culture broth were centrifuged for one minute at 12,000rpm. After discarding the supernatant, strains were fixed in a Karnovsky solution (2.5% glutaraldehyde in phosphate buffer 0.1M [pH 7.3]) for four hours. Samples were removed from the fixer and washed three times for five minutes in distilled water. Next the samples were immersed in 0.5% osmium tetroxide for 40min, before being washed three times for 10min in distilled water. Samples were dehydrated using increasing concentrations of alcohol: two times for 10min in 7.5% alcohol; two times for 10min in 15% alcohol; two times for 10min in 30% alcohol; two times for 10min in 50% alcohol; three times for 15min in 70% alcohol; two times for 15min in 90% alcohol; two times for 10min in 100% alcohol. After the dehydration the stubs were mounted and samples metalized. The analyses were performed in an electronic microscopy Tecnai Spirit Fei Company and the images were obtained at a magnification of 30000×.
ResultsIdentification of isolatesThe identification of CoNS through the biochemical tests detected 223 (74.3%) S. epidermidis, 27 (9.0%) S. hemolyticus, 22 (7.3%) S. hominis, 14 (4.7%) S. warneri, 9 (3.0%) S. lugdunensis, and 5 (1.7%) S. capitis. Through the ITS-PCR method, 223 (74.3%) S. epidermidis, 29 (9.7%) S. hemolyticus, 23 (7.7%) S. hominis, 11 (3.7%) S. warneri, 9 (3.0%) S. lugdunensis, and 5 (1.7%) S. capitis were detected. Agreement between the identification methods was found in 98% of cases.
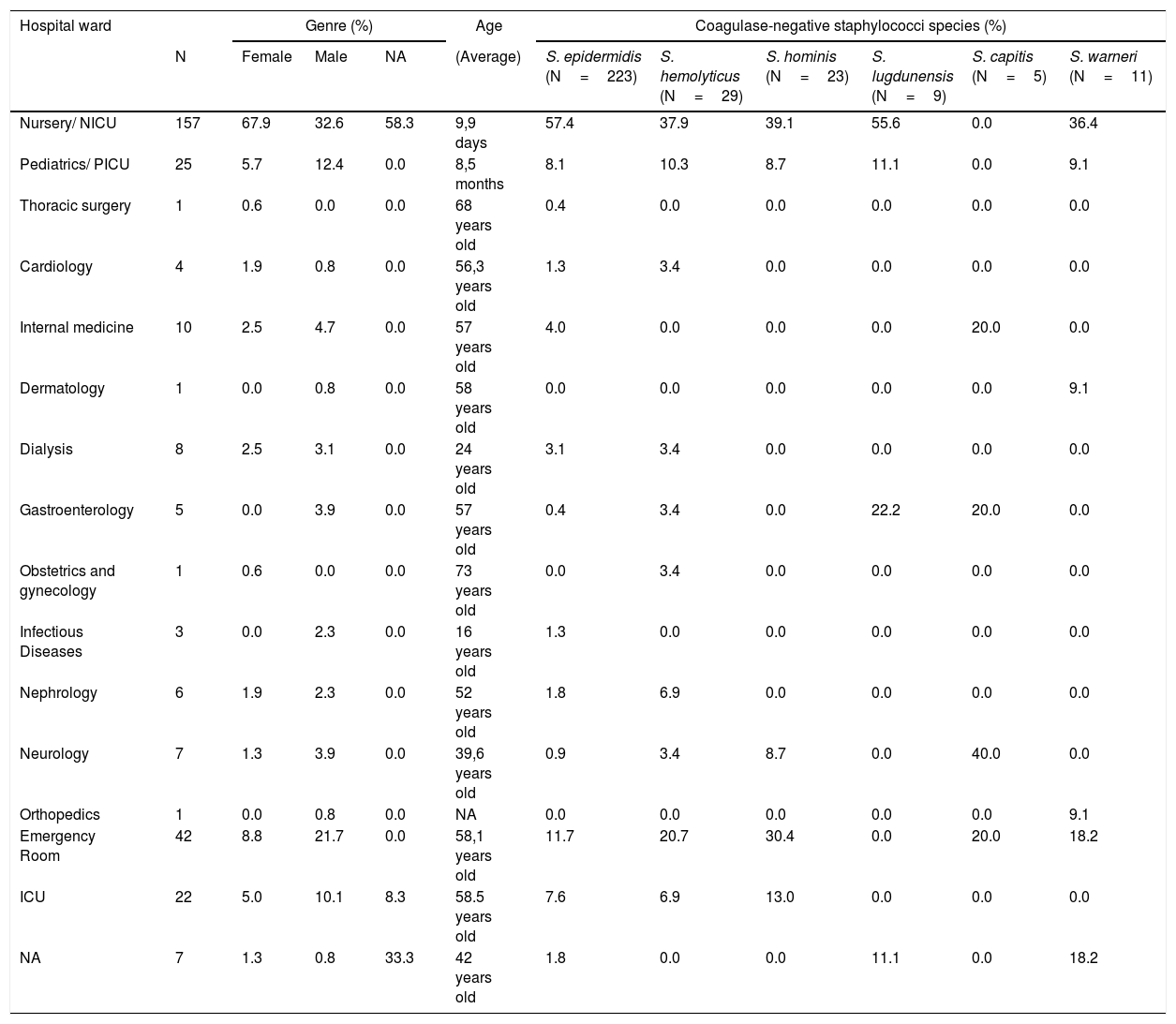
Table 1 presents the source of the CoNS species isolates, as well as the demographic data of the patients with bacteremia.
Origin of CoNS isolates and demographic data of patients with bacteremia at Hospital das Clínicas de Botucatu - Paulista State University between 1990 and 2009.
| Hospital ward | Genre (%) | Age | Coagulase-negative staphylococci species (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Female | Male | NA | (Average) | S. epidermidis (N=223) | S. hemolyticus (N=29) | S. hominis (N=23) | S. lugdunensis (N=9) | S. capitis (N=5) | S. warneri (N=11) | |
| Nursery/ NICU | 157 | 67.9 | 32.6 | 58.3 | 9,9 days | 57.4 | 37.9 | 39.1 | 55.6 | 0.0 | 36.4 |
| Pediatrics/ PICU | 25 | 5.7 | 12.4 | 0.0 | 8,5 months | 8.1 | 10.3 | 8.7 | 11.1 | 0.0 | 9.1 |
| Thoracic surgery | 1 | 0.6 | 0.0 | 0.0 | 68 years old | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Cardiology | 4 | 1.9 | 0.8 | 0.0 | 56,3 years old | 1.3 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 |
| Internal medicine | 10 | 2.5 | 4.7 | 0.0 | 57 years old | 4.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 |
| Dermatology | 1 | 0.0 | 0.8 | 0.0 | 58 years old | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 |
| Dialysis | 8 | 2.5 | 3.1 | 0.0 | 24 years old | 3.1 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 |
| Gastroenterology | 5 | 0.0 | 3.9 | 0.0 | 57 years old | 0.4 | 3.4 | 0.0 | 22.2 | 20.0 | 0.0 |
| Obstetrics and gynecology | 1 | 0.6 | 0.0 | 0.0 | 73 years old | 0.0 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 |
| Infectious Diseases | 3 | 0.0 | 2.3 | 0.0 | 16 years old | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Nephrology | 6 | 1.9 | 2.3 | 0.0 | 52 years old | 1.8 | 6.9 | 0.0 | 0.0 | 0.0 | 0.0 |
| Neurology | 7 | 1.3 | 3.9 | 0.0 | 39,6 years old | 0.9 | 3.4 | 8.7 | 0.0 | 40.0 | 0.0 |
| Orthopedics | 1 | 0.0 | 0.8 | 0.0 | NA | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 |
| Emergency Room | 42 | 8.8 | 21.7 | 0.0 | 58,1 years old | 11.7 | 20.7 | 30.4 | 0.0 | 20.0 | 18.2 |
| ICU | 22 | 5.0 | 10.1 | 8.3 | 58.5 years old | 7.6 | 6.9 | 13.0 | 0.0 | 0.0 | 0.0 |
| NA | 7 | 1.3 | 0.8 | 33.3 | 42 years old | 1.8 | 0.0 | 0.0 | 11.1 | 0.0 | 18.2 |
N, number; NICU, neonatal intensive care unit; PICU, pediatrics intensive care unit; ICU, intensive care unit; NA, not available.
The mecA gene was detected in 222 (74%) of the studied samples, found in 78.5% of S. epidermidis, 75.9% of S. hemolyticus, 69.6% of S. hominis, 27.3% of S. warneri, 80% of S. capitis, and 22.2% of S. lugdunensis.
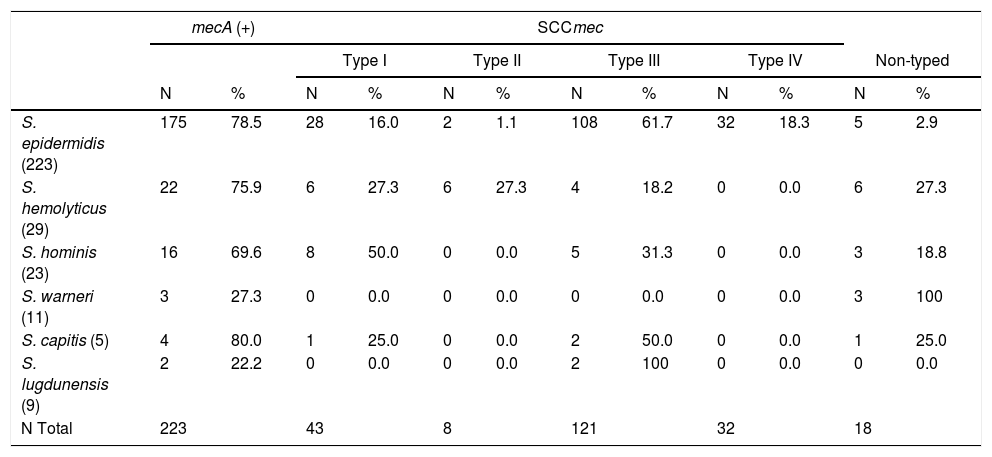
The characterization of SCCmec in mecA-positive strains was as follows: 43 (19.4%) were classified as type I, 8 (3.6%) as type II, 121 (54.5%) as type III, 32 (14.4%) as type IV, and 18 (8.1%) were not typed by this method. The correlations between the SCCmec types and methicillin-resistant CoNS species are presented in Table 2.
Classification of SCCmec types among the methicillin-resistant CoNS species.
| mecA (+) | SCCmec | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type I | Type II | Type III | Type IV | Non-typed | ||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | |
| S. epidermidis (223) | 175 | 78.5 | 28 | 16.0 | 2 | 1.1 | 108 | 61.7 | 32 | 18.3 | 5 | 2.9 |
| S. hemolyticus (29) | 22 | 75.9 | 6 | 27.3 | 6 | 27.3 | 4 | 18.2 | 0 | 0.0 | 6 | 27.3 |
| S. hominis (23) | 16 | 69.6 | 8 | 50.0 | 0 | 0.0 | 5 | 31.3 | 0 | 0.0 | 3 | 18.8 |
| S. warneri (11) | 3 | 27.3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 3 | 100 |
| S. capitis (5) | 4 | 80.0 | 1 | 25.0 | 0 | 0.0 | 2 | 50.0 | 0 | 0.0 | 1 | 25.0 |
| S. lugdunensis (9) | 2 | 22.2 | 0 | 0.0 | 0 | 0.0 | 2 | 100 | 0 | 0.0 | 0 | 0.0 |
| N Total | 223 | 43 | 8 | 121 | 32 | 18 | ||||||
N, number of samples.
The MICs of the antimicrobials used for the treatment of CoNS infections was determined by the E-test. Oxacillin resistance was found in 206 (68.7%) CoNS strains and 25 were tigecycline resistant (8.3%). Resistance to tigecycline was detected in 4.5% of S. epidermidis, 13.8% of S. hemolyticus, 13% of S. hominis, and 11.1% of S. lugdunensis. For quinupristin/dalfopristin, one S. epidermidis isolate presented intermediate resistance with an MIC of 2μg/mL, and one S. epidermidis and one S. hemolyticus with MICs of 3μg/mL. The sensitivity and specificity of the oxacillin MIC by the E-test compared with the presence of mecA were, respectively, 87.4% and 82.3%.
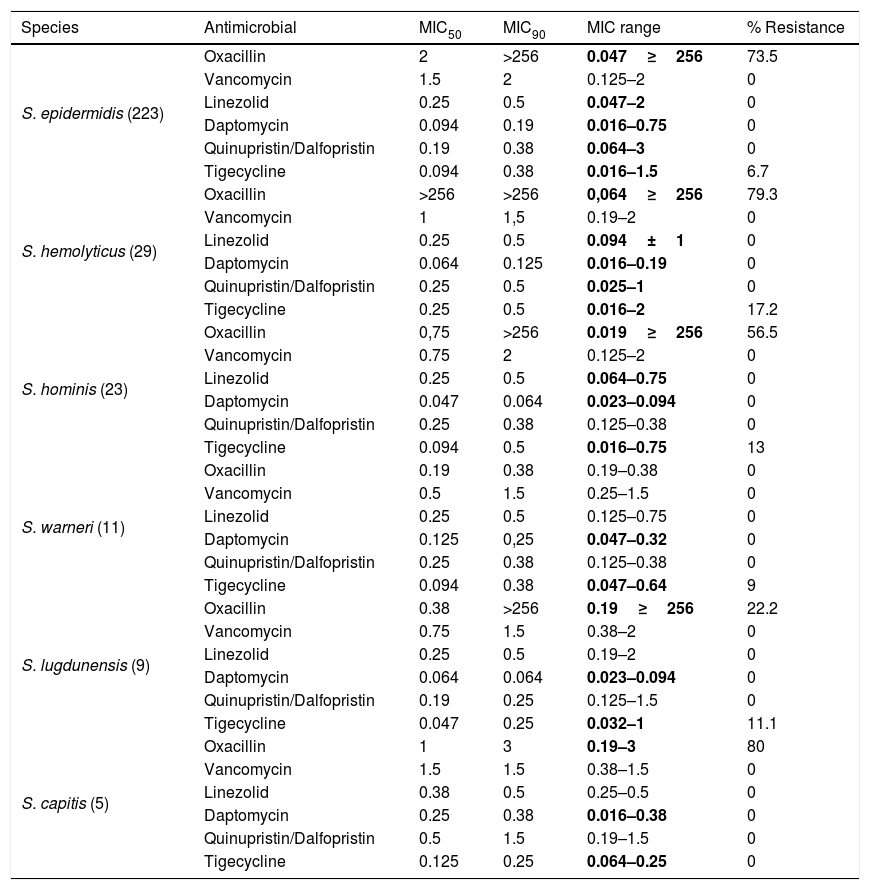
The antimicrobial MICs for inhibition of 50% and 90% of strains (MIC50 and MIC90) were, respectively, 2μL/mL and >256μL/mL for oxacillin, 1.5μL/mL and 2μL/mL for vancomycin, 0.25μL/mL and 0.5μL/mL for linezolid, 0.094μL/mL and 0.19μL/mL for daptomycin, 0.19μL/mL and 0.5μL/mL for quinupristin/dalfopristin, and 0.125μL/mL and 0.38μL/mL for tigecycline.
With respect to CoNS species, the parameter distribution of MIC50 and MIC90 revealed high rates of oxacillin resistance in S. hemolyticus. This was the only CoNS species for which the minimal concentration to inhibit 50% of isolates was >256μg/mL, reflecting the high resistance rate of S. hemolyticus to oxacillin (Table 3).
Determination of MIC50, MIC90, MIC range (μg/ml), and antimicrobial resistance in CoNS species.
| Species | Antimicrobial | MIC50 | MIC90 | MIC range | % Resistance |
|---|---|---|---|---|---|
| S. epidermidis (223) | Oxacillin | 2 | >256 | 0.047≥256 | 73.5 |
| Vancomycin | 1.5 | 2 | 0.125–2 | 0 | |
| Linezolid | 0.25 | 0.5 | 0.047–2 | 0 | |
| Daptomycin | 0.094 | 0.19 | 0.016–0.75 | 0 | |
| Quinupristin/Dalfopristin | 0.19 | 0.38 | 0.064–3 | 0 | |
| Tigecycline | 0.094 | 0.38 | 0.016–1.5 | 6.7 | |
| S. hemolyticus (29) | Oxacillin | >256 | >256 | 0,064≥256 | 79.3 |
| Vancomycin | 1 | 1,5 | 0.19–2 | 0 | |
| Linezolid | 0.25 | 0.5 | 0.094±1 | 0 | |
| Daptomycin | 0.064 | 0.125 | 0.016–0.19 | 0 | |
| Quinupristin/Dalfopristin | 0.25 | 0.5 | 0.025–1 | 0 | |
| Tigecycline | 0.25 | 0.5 | 0.016–2 | 17.2 | |
| S. hominis (23) | Oxacillin | 0,75 | >256 | 0.019≥256 | 56.5 |
| Vancomycin | 0.75 | 2 | 0.125–2 | 0 | |
| Linezolid | 0.25 | 0.5 | 0.064–0.75 | 0 | |
| Daptomycin | 0.047 | 0.064 | 0.023–0.094 | 0 | |
| Quinupristin/Dalfopristin | 0.25 | 0.38 | 0.125–0.38 | 0 | |
| Tigecycline | 0.094 | 0.5 | 0.016–0.75 | 13 | |
| S. warneri (11) | Oxacillin | 0.19 | 0.38 | 0.19–0.38 | 0 |
| Vancomycin | 0.5 | 1.5 | 0.25–1.5 | 0 | |
| Linezolid | 0.25 | 0.5 | 0.125–0.75 | 0 | |
| Daptomycin | 0.125 | 0,25 | 0.047–0.32 | 0 | |
| Quinupristin/Dalfopristin | 0.25 | 0.38 | 0.125–0.38 | 0 | |
| Tigecycline | 0.094 | 0.38 | 0.047–0.64 | 9 | |
| S. lugdunensis (9) | Oxacillin | 0.38 | >256 | 0.19≥256 | 22.2 |
| Vancomycin | 0.75 | 1.5 | 0.38–2 | 0 | |
| Linezolid | 0.25 | 0.5 | 0.19–2 | 0 | |
| Daptomycin | 0.064 | 0.064 | 0.023–0.094 | 0 | |
| Quinupristin/Dalfopristin | 0.19 | 0.25 | 0.125–1.5 | 0 | |
| Tigecycline | 0.047 | 0.25 | 0.032–1 | 11.1 | |
| S. capitis (5) | Oxacillin | 1 | 3 | 0.19–3 | 80 |
| Vancomycin | 1.5 | 1.5 | 0.38–1.5 | 0 | |
| Linezolid | 0.38 | 0.5 | 0.25–0.5 | 0 | |
| Daptomycin | 0.25 | 0.38 | 0.016–0.38 | 0 | |
| Quinupristin/Dalfopristin | 0.5 | 1.5 | 0.19–1.5 | 0 | |
| Tigecycline | 0.125 | 0.25 | 0.064–0.25 | 0 |
MIC, Minimal Inhibitory Concentration. MIC50, Minimal Concentration necessary to inhibit 50% of bacterial growth. MIC90, Minimal Concentration necessary to inhibit 90% of bacterial growth.
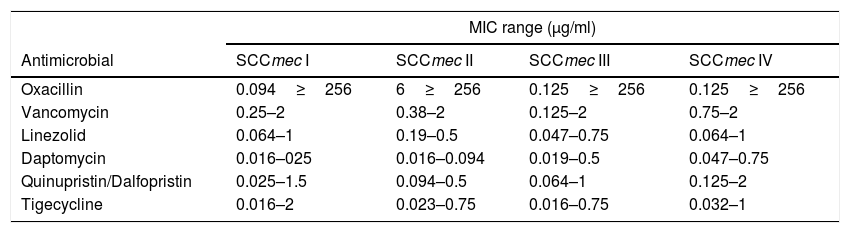
The MIC range of each antimicrobial was compared to the SCCmec type. The isolates typed as SCCmecI presented higher tigecycline MICs and those classified as SCCmecII presented the highest oxacillin MICs. The SCCmecIV strains showed the highest MIC values for vancomycin, linezolid, daptomycin, and quinupristin/dalfopristin (Table 4).
Determination of the MIC range according to SCCmec.
| MIC range (μg/ml) | ||||
|---|---|---|---|---|
| Antimicrobial | SCCmec I | SCCmec II | SCCmec III | SCCmec IV |
| Oxacillin | 0.094≥256 | 6≥256 | 0.125≥256 | 0.125≥256 |
| Vancomycin | 0.25–2 | 0.38–2 | 0.125–2 | 0.75–2 |
| Linezolid | 0.064–1 | 0.19–0.5 | 0.047–0.75 | 0.064–1 |
| Daptomycin | 0.016–025 | 0.016–0.094 | 0.019–0.5 | 0.047–0.75 |
| Quinupristin/Dalfopristin | 0.025–1.5 | 0.094–0.5 | 0.064–1 | 0.125–2 |
| Tigecycline | 0.016–2 | 0.023–0.75 | 0.016–0.75 | 0.032–1 |
MIC, Minimal Inhibitory Concentration.
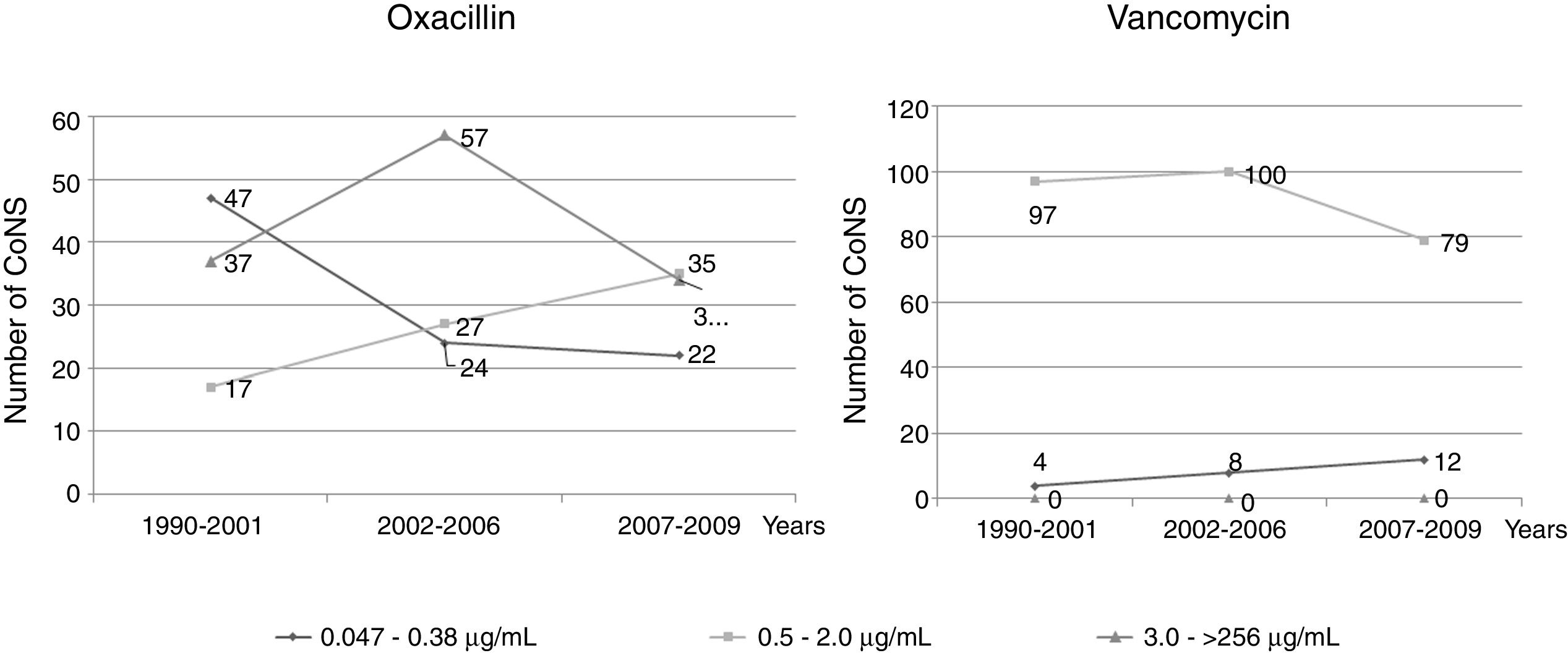
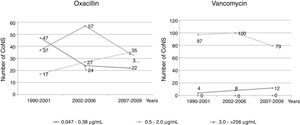
An evaluation was performed of the oxacillin and vancomycin MICs over a period of 20 years, divided into three time periods according to the distribution of oxacillin and vancomycin MIC ranges. For oxacillin, in the first period (1990–2001), higher values of MIC were observed, but still indicating susceptibility. In the other periods (2002–2006 and 2007–2009), a decrease in the MIC values was observed, still indicating susceptibility, as well as an increase in the resistance rates. Regarding vancomycin, the same evaluation demonstrated significant differences in the three periods among strains with MICs from 0.047μg/mL to 0.38μg/mL and from 0.5μg/mL to 2.0μg/mL (Fig. 1).
Determination of the reduced susceptibility to vancomycinA growth of 214 (71.3%) strains was observed on the BHI plate with 4μg/mL of vancomycin, 89 (29.7%) on the plate with 6μg/mL of vancomycin, and 8 (2.6%) isolates on the 8μg/mL vancomycin plate. None of the isolates grew in the medium with 16μg/mL of vancomycin. There were no significant differences in MICs between strains that presented no growth on the plate complemented with vancomycin and those which grew on the 4μg/mL vancomycin plate. Among those that showed growth on the medium with 6μg/mL of vancomycin, the MIC range was 0.5–2μg/mL. Among those which grew on 8μg/mL of vancomycin, the MIC range was 0.75–24μg/mL. The strains grown on 8μg/mL of vancomycin were four S. epidermidis, one S. hemolyticus, two S. hominis, and one S. capitis, all resistant to oxacillin, and S. epidermidis and S. capitis being carriers of SCCmec III, S. hominis of SCCmec I, and S. hemolyticus with non-typed SCCmec.
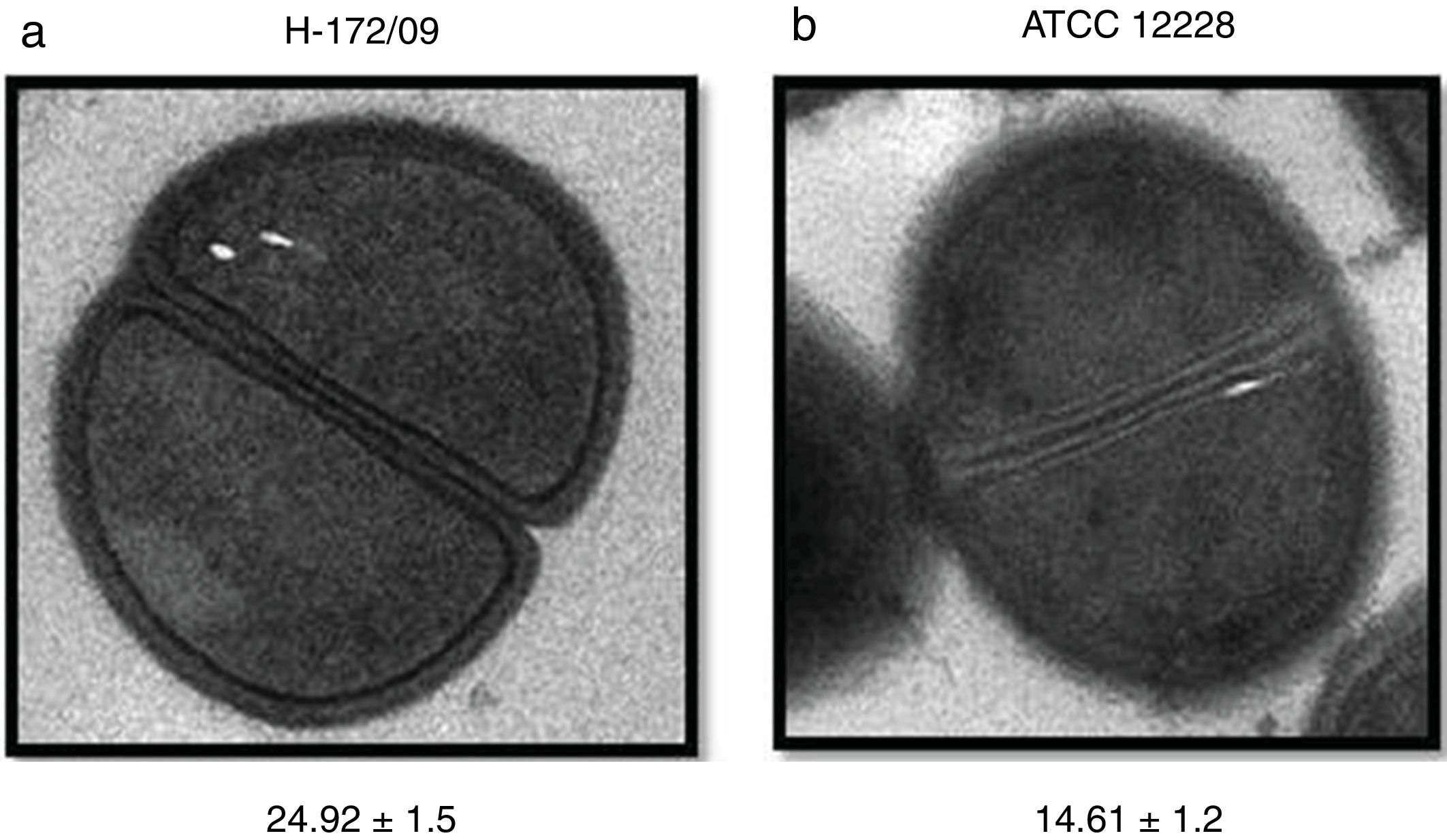
Analysis of the cell wall thickness of strains with reduced susceptibility to vancomycinThe cell wall thickness under cell division was measured in eight strains (values are presented in nanometers, mean±SD). The four S. epidermidis strains presented mean values of 21.66±1.4, 20.12±0.9, 24.24±1.3, and 14.20±1.3nm. The S. hemolyticus isolate showed a cell wall thickness of 24.88±1.7nm. For the S. capitis isolate, the cell wall thickness was 19.33±1.8, and for the two S. hominis isolates the values were 17.68±2.1 and 24.92±1.5. For comparison, the used reference strains S. epidermidis ATCC 12228 and S. hemolyticus ATCC29970 showed cell wall thicknesses of, respectively, 14.61±1.2 and 14.73±0.7nm. The comparison of the studied strains against controls showed higher values, ranging from 17nm to 24.92nm, with the exception of one S. epidermidis isolate, which presented a lower value compared to the controls (Fig. 2).
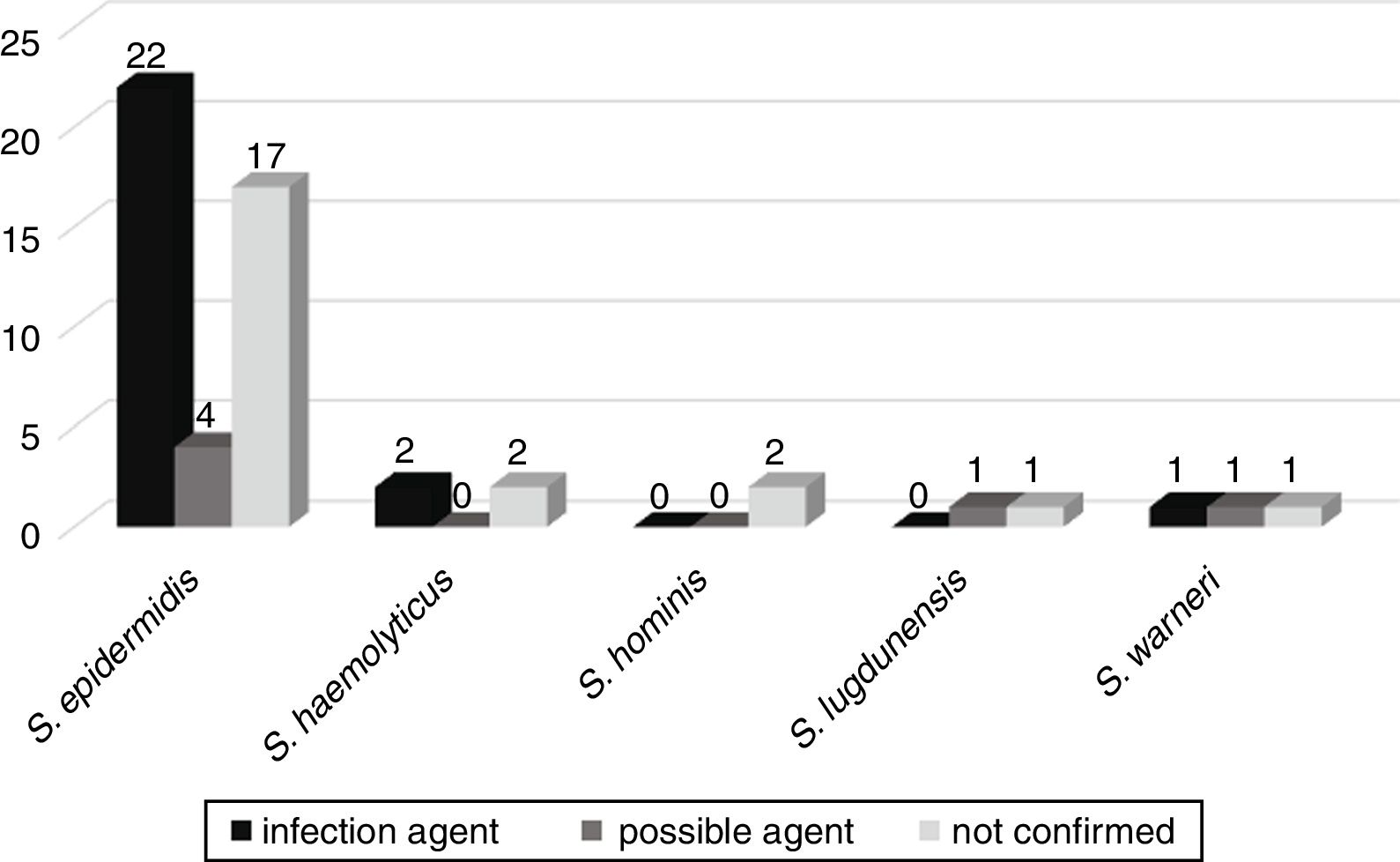
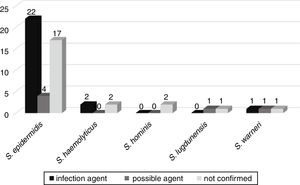
CoNS bacteremia evolutionThe possible evolution of bacteremia to other infections, such as sepsis, pneumonia, urinary tract infection, meningitis, peritonitis, necrotizing enterocolitis, omphalitis, and brain abscess, was followed up in 52 patients admitted to the neonatal wards. Of these, in 25 (46.3%) patients the same CoNS isolated from blood culture was confirmed to be the etiologic agent of infection. In six (11.1%), CoNS was the possible agent of infection, and in 23 (42.6%) CoNS was not related to the patient's infection (Fig. 3).
The mecA gene was detected in 33 (61.1%) of these CoNS, with 20 (60.6%) CoNS associated with more severe infections or possible agents of these infections.
DiscussionThe CoNS are considered one of the main causes of bacteremia. The importance of these bacteria has increased in the hospital environment during recent years, mostly due to antimicrobial resistance. In the present work, 300 CoNS strains isolated from blood cultures of inpatients at the Hospital of Clinics of Botucatu, over a period of 20 years, were studied. These isolates were characterized regarding their antimicrobial susceptibility.
The mecA detection determined oxacillin resistance in 78.5% of CoNS, and the MIC50 value (2μg/mL) was indicative of resistance to this drug, confirming the low susceptibility of these bacteria to β–lactam agents. Previous works have demonstrated that since the 1970s the CoNS isolates have presented higher oxacillin resistance rates than S. aureus.14,15 The β–lactam resistance rates have been shown to range from 65% to 95% in hospitals in Brazil and in other countries.2,16,17 The highest rates of oxacillin resistance were found in S. epidermidis, followed by S. hemolyticus, S. hominis, and S. capitis. Similar results were described in previous studies, which showed oxacillin resistance in 97% of S. epidermidis between 1999 and 2001 in a neonatal intensive care unit (NICU),18 in 96% of S. hemolyticus isolated in Brazil,19 and in 100% of S. hominis isolated in an NICU from Spain.20 Regarding S. capitis, discrepant results were found in the studies conducted by Caierão et al.21
Although at a lower frequency, the mecA gene was detected in S. warneri and S. lugdunensis. S. warneri has been described with rates of 33.3% of oxacillin resistance in NICUs,22 and the first description of mecA in S. lugdunensis was in the study conducted by Kawaguchi et al.23 Despite the low resistance rates,24 determination of antimicrobial susceptibility of S. lugdunensis is important, not only due to its clinical implications, since this species is the most aggressive of the CoNS, but also for the establishment of early treatment with adequate antimicrobials and good clinical results.25
The characterization of the SCCmec showed the presence of types I-IV in the isolates, with SCCmecIII being the most frequently detected in the studied strains, mainly in S. epidermidis, the only species that carried type IV. SCCmec type III is the largest of them all, codifies for several resistance-associated genes, and is the most commonly isolated CoNS from hospital specimens. S. epidermidis is the main colonizer of the human skin and the most commonly detected in infection sites. The selective pressure in the hospital environment leads to dissemination of SCCmec III strains, which are associated with serious infections.26 SCCmec type IV has been related with community-associated Staphylococcus spp., and was described for the first time in an S. epidermidis strain, a fact that implies its transference from S. epidermidis to S. aureus. A reduced cost transfer of SCCmec IV due to its small size would probably lead to a higher incidence of infections caused by SCCmec IV carriers.12,27 SCCmecII presented an association with S. hemolyticus, similar to the findings of Machado et al.,12 whose work only detected type II in this species.
The alternative for the treatment of oxacillin resistant staphylococci is vancomycin, a glycopeptide first used in 1958 in invasive infections. There are, however, descriptions of resistance and reduced susceptibility to this drug,6 although unstable.28 The results of the current work showed that, with the exception of oxacillin, vancomycin was the drug with the highest values of MIC50 and MIC90. Despite the full susceptibility of the collection, the MIC values were the highest in the period of 20 years, and reduced susceptibility was detected in some S. epidermidis, S. hemolyticus, S. hominis, and S. capitis strains. In the studies performed by Natoli et al.,29 reduced susceptibility to vancomycin was detected with a frequency of 5.4% among CoNS isolates, in S. epidermidis and S. hemolyticus species. According to the same authors, Staphylococcus colonies grown on vancomycin agar may lead to glycopeptide heteroresistance, which can be a precursor of glycopeptide resistance, causing complicated infections and treatment failure. This should be taken into consideration in therapeutic decisions.30
The increase in the proportion of oxacillin resistant CoNS and decrease in their susceptibility to vancomycin emphasize the importance of studies involving other therapeutic choices. Tigecycline, a Gram-positive and negative broad spectrum semisynthetic glycylcycline, is considered a drug with excellent activity against oxacillin susceptible and resistant CoNS,14,29 despite our data showing rates of 8.3% of resistance, most often in S. hemolyticus, followed by S. hominis, S. lugdunensis, and S. epidermidis. Similar MIC90 results were described for tigecycline, with no difference between oxacillin resistant and susceptible CoNS.31 In the studies conducted by Natoli et al.29 tigecycline demonstrated good activity against CoNS. According to those authors, tigecycline is not recommended for the treatment of bacteremia and its use should be limited in order to preserve activity against multi-resistant Gram-negative bacteria. Mutations in the ribosomal gene S10 and rpsJ and mepA genes seem to be associated with tigecycline resistance.32,33
Quinupristin/dalfopristin is a streptogramin belonging to the macrolide-lincosamide-streptogramin group. The combination of quinupristin and dalfopristin is synergistic and usually bactericidal when these agents are compared singly, or compared to similar macrolide antimicrobials.34 In this work, despite presenting excellent efficacy in the majority of the studied CoNS, intermediate resistance to this drug was detected in two S. epidermidis and one S. hemolyticus. In the study conducted by Mendes et al.,35 quinupristin/dalfopristin demonstrated excellent effectiveness for CoNS, the strains were fully susceptible, with MIC90 values of 0.38μg/mL for the oxacillin-susceptible strains and 0.75μg/mL for the oxacillin resistant strains. In addition, Venkatesh et al.36 described good effectiveness of quinupristin/dalfopristin for oxacillin resistant and susceptible strains.
Daptomycin, an antimicrobial studied for decades, was brought back into use in 2006, for the treatment of bacteremia and endocarditis caused by Staphylococcus.37 In the present study, daptomycin showed excellent efficacy for CoNS isolates, with low MIC50 values. Olivares et al.37 also verified the susceptibility of all CoNS to daptomycin, with low variation in the MIC values. In a work studying 1126 CoNS, conducted by Critchley et al.,38 daptomycin was active on a MIC range from 0.015 to 2.0μg/mL, and the MIC90 was 0.5μg/mL. According to those authors, the most active agents in these analyses were daptomycin and quinupristin/dalfopristin, which emphasizes the relevance of these antimicrobials in the treatment of infections caused by CoNS.
Linezolid, a synthetic oxazolidinone potentially active against several bacteria, is another drug that has demonstrated good efficacy against CoNS.39 Linezolid has become important for the therapeutic treatment of chronic infections by CoNS, despite the description of resistance. In Brazil, the first case of linezolid resistance was described in 2006, on a clinical isolate of MRSA40 and, among the CoNS, in S. epidermidis, S. hominis, and S. hemolyticus.41 In the work conducted by Olivares et al.,37 despite the detection of seven linezolid resistant strains, this drug presented effective MIC values in CoNS strains. On the other hand, an outbreak of linezolid resistant S. epidermidis clones containing mutations in ribosomal proteins L3 and L4, as well as the cfr plasmid, recently identified in France, have been reported.42
S. hemolyticus was the species that presented the highest oxacillin MIC values, and concentrations higher than 256μg/mL were needed to inhibit 50% of isolates. As well as for oxacillin, this species presented higher MIC values for tigecycline and most of the antimicrobials tested. Several works show high rates of resistance to the antimicrobials used for the treatment of infections caused by S. hemolyticus, especially methicillin and glycopeptides. Some authors describe a high prevalence of genes encoding resistance to β-lactam and aminoglycoside agents, as well as a significant proportion of isolates with MIC values close to the resistance breakpoint,43,44 emphasizing the importance of S. hemolyticus as a multiresistant pathogen.
The association of the MIC range with the SCCmec types demonstrated higher MICs for vancomycin, linezolid, daptomycin, and quinupristin/dalfopristin in S. epidermidis carrying SCCmecIV. The genes related to resistance to these antimicrobials are mediated by plasmids, found mostly in methicillin resistant strains with hospital origin.45 SCCmec IV is characteristic of community isolates, especially due to its size and low adaptive cost. Studies suggest that the acquisition of resistance genes has environmental non-clinical origin, given the high diversity of resistance gene carriers in the natural environment.46,47 Furthermore, SCCmecIV was only detected in S. epidermidis, which as the most common species in human skin is the most influenced by selective pressure. The higher MICs compared to other antimicrobials may also be related to the selective pressure, as the level of exposure to these drugs in the hospital environment would bias the selection of reduced susceptibility and resistant strains, since several mutation events, genetic recombination, and modifications in the microbial physiology are needed to generate phenotypic changes.48
Metabolic and physiological modifications related to selective pressure could be observed in strains grown on 8μg/mL vancomycin agar, as the cell wall thickening was found in most CoNS isolates due to increased peptidoglycan synthesis. This mechanism is more advantageous for CoNS than the acquisition of the van operon, which is a mediator of vancomycin resistance. The van operon is acquired by horizontal gene transfer and its expression is only stimulated in the presence of glycopeptide. This stimulus causes a very high adaptive cost in the presence of vancomycin, being disadvantageous for the CoNS in a vancomycin medium.48,49 Regarding the acquisition of other resistance genes in CoNS, such as the mecA gene, the initial adaptive cost is softened by compensatory additional mutations for the resistance “costs”. Recently, punctual mutations in genes such as vraR have been shown to be associated with reduced susceptibility to vancomycin and cell wall thickening in staphylococci.50
The present study showed a high rate of bacteremia caused by CoNS in neonatal and pediatric units, involving children under one year of age, the main agents being S. epidermidis, S. hemolyticus, S. hominis, S. lugdunensis, and S. warneri. S. capitis were not isolated in these units, being agents of bacteremia in adults admitted to the internal medicine ward, gastroenterology, neurology, and emergency room.
As the study involved bacteremia caused by staphylococci isolated over more than 30 years, much of the data associated with patients’ medical records could not be retrieved, so it was not possible to associate the infection outcome of all patients, which is a limitation of the current study. Data obtained from patients in neonatal units indicated the clinical importance of CoNS bacteremia, which may progress to diseases that require specialized care.
Knowledge of antimicrobial resistance is of great importance for the correct treatment of infections caused by CoNS. Furthermore, the virulence factors of these bacteria and the immunity of patients are factors that contribute to the ability of the microorganism to cause more serious infections.
The results of our study spanning 20 years showed a high frequency of antimicrobial resistance in CoNS, which reflects the excessive use of these drugs. Besides the metabolic and physiologic modifications that could lead to reduced susceptibility to antimicrobials, their condition as human commensal bacteria make them ideal transporters and an efficient reservoir of resistance genes, especially the low-cost elements, such as SCCmec.45 The antimicrobials which offered the best results should be used in such a way as to preserve their efficacy and prevent resistance.
Conflicts of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the São Paulo Research Foundation (FAPESP- Process: 2011/23742-2)) and the National Council for Scientific and Technological Development (CNPq- Process: 470649/2011-9) for the financial support.