The frequencies of the Human leukocyte antigen (HLA) alleles in the Puyanawa indigenous reserve population and their association with the NDO-LID and ELISA PGL-1 rapid serological test was assessed. This was a cross-sectional study with an epidemiological clinical design conducted in two indigenous communities in the state of Acre, Brazil. Blood was collected in a tube with EDTA to identify HLA alleles and perform serological tests. DNA was obtained using the salting out procedure. The LabType™ technique (One-Lambda-USA) was used for HLA class I (loci A*, B* and C*) and II (loci DRB1*, DQA1* and DQB1*) typing. Allele frequency was obtained by direct count, and the chi-square test was used to assess the association with the NDO-LID and PGL-1 tests. The most frequent alleles in the two communities were: HLA-A*02:01, HLA-B*40:02, HLA-DRB1*16:02, HLA-DQA1*05:05 and HLA-DQB1*03:01. The allele HLA-C*04:01 was the most common in the Barão community, and the allele HLA-C*07:01 in Ipiranga. Among individuals who presented seropositivity to the NDO-LID test, the association with alleles HLA-A*02 (43.18% vs 24.8%, p=0.03, OR=2.35) and HLA-B*53 (6.83% vs 0.0%, p=0.03, OR=8.95) was observed in the Barão community. HLA-B*15 was associated with non-seroconversion to the NDO-LID test in Ipiranga. In both communities, HLA-B*40 and HLA-C*03 were associated with positive serological response to ELISA PGL-1. The HLA class I and II alleles most frequently found in this study have already been described among Terena indigenous groups, and HLA class I contributes to seroconversion to NDO-LID and PGL-1 tests in inhabitants of the Barão and Ipiranga communities.
Leprosy is a chronic granulomatous infection caused by the Mycobacterium leprae (M. leprae)1 that has affected humanity for centuries. Even after the introduction of polychemotherapy, it remains a public health problem in Brazil.2 The infection is transmitted mainly among family members who share genetic and environmental factors, besides being considerably influenced by the variation in genes related to immunity.3–7
Many diseases – including leprosy – have been related to the HLA system due to its importance in adaptive immune response.4,8–10 Studies designed as family aggregation, complex segregation, linkage, association, and genome scans suggest that leprosy phenotypes largely depend on the host's genetic characteristics. Numerous positive associations have linked class I and II molecules with susceptibility to leprosy, its clinical forms and, leprosy reactions.3,11–14
An experimental study conducted with Vietnamese and Indian individuals showed a strong association of HLA-C (HLA-C*15:05) in the susceptibility to leprosy.11 The presence of HLA-A*28, in Mexico's mixed-race population, has been defined as a predisposition marker to lepromatous leprosy (LL).13 In Brazil, a positive association was found between HLA-A*11, HLA-B*38, HLA-C*12, and leprosy,12 and alleles A*03 and A*11 were associated with susceptibility to develop borderline leprosy (BL).3 The protective association was observed between clinical borderline leprosy and HLA-C*05.6
Regarding class II molecules, studies indicate a more consistent association between HLA-DRB1/DQB1 alleles and susceptibility to leprosy, suggesting that these could be the main genes associated with leprosy susceptibility.8–10 Several studies have reported association of alleles HLA-DRB1*04, DRB1*10, DRB1*12, DRB1*15 and DRB1*16 with susceptibility or resistance to leprosy in several populations worldwide.8–10,14 Studies show that alleles HLA-DRB1*15, and DRB1*16 are associated with leprosy, especially in Indian, Japanese, and Brazilian populations,5,8,9,15,16 whereas alleles HLA-DRB1*04 and HLA-DRB1*07 are associated to protection,5,17 and the latter with BL.6
Serological methods are indicators of the spread of the M. leprae infection in the general population and can contribute positively as an epidemiological surveillance instrument of leprosy.18–20 These methods use techniques based on the detection of specific anti-M. leprae antibodies. Thus, those with positive responses to the tests have higher diversity of antibodies against the bacillus, a response presented mainly in multibacillary cases (MB).18,21,22
Seropositivity to phenolic glycolipid 1 (PGL-1) estimates a three-fold greater chance of developing the disease.23,24 Wambier et al.,25 observed that the PGL-1 seropositivity rate was affected by kinship, being higher among siblings, followed by parents, spouses, and children, suggesting that transmission of the disease occurs mainly among individuals who share genetic and environmental aspects.
LID-1 (Leprosy IDRI Diagnostic), derived from the fusion between recombinational proteins ML0405 and ML2331 and the PGL-1 antigen, represent important antigens specific to leprosy serology, and studies have suggested that their association improves serological sensitivity to diagnose paucibacillary leprosy (PB) and early disease.26–28 Such good serological reactivity led to the development of a simple and rapid test incorporating the LID-1 and PGL-1 antigens, immobilized in a nitrocellulose membrane for the detection of antibodies, aiding in the leprosy diagnosis.27
The state of Acre, an endemic region for leprosy in Brazil, maintains a high detection coefficient of 15.07 per 100 thousand inhabitants.29
The occurrence of MB cases in Ipiranga community, the possible detection failure in early disease,30 contact with the regional population, and cultural and socioeconomic habits of indigenous peoples, are substrates that predispose to a higher risk and exposure to the disease, justifying an investigation.
To analyze genetic and serological characteristics in an indigenous community is not a common and easy task. Given this context, this study was an opportunity to gather knowledge about leprosy, HLA genotypes, and the contribution of these molecules to seroconversion to NDO-LID and ELISA PGL-1 tests in this never-studied ethnic group.
Thus, assuming the participation of HLA alleles in the immunological response in leprosy and the importance of serology in the identification of subclinical infection and as a diagnostic aid, this study proposes to identify the frequencies of HLA alleles in the indigenous population of the Puyanawa reserve, and to assess their association with the results of the rapid lateral flow serological test NDO-LID and ELISA PGL-1.
Material and methodThis was a cross-sectional study with epidemiological clinical design conducted in two indigenous communities, Barão and Ipiranga, in the municipality of Mâncio Lima, Acre, Brazil. These communities are part of the territory of the indigenous ethnic group Puyanawa with estimated population of 576 individuals – 320 in Barão and 256 in Ipiranga, according to data from the Indigenous Health Care Information System.
The sample consisted of 284 individuals, over 18 years of age, who had lived for more than one year in the communities without prior diagnosis of leprosy. Among the studied individuals, 145 (51%) were men, mean age 36, ranging from 18 to 31 years.
SamplesTo identify HLA alleles and conduct the serological tests, 10mL peripheral venous blood was collected in a tube with ethylenediaminetetraacetic acid (EDTA). Of the obtained material, 5mL were used to extract plasma, which was separated into aliquots and stored at −20°C. The rest of the material (5mL) was stored at 2–8°C. One aliquot was used to perform the NDO-LID test, and the remaining sample was sent to the Lauro de Souza Lima Institute in Bauru/SP for identification of the HLA alleles and detection of ELISA PGL-1.
Serological testsDetection of IgM antibodies against PGL-1 of M. leprae was performed by ELISA technique, developed according to the methodology described by Brett et al., 1983. All sera were tested in duplicate and the ELISA results were considered positive when optical density (OD) was equal to or greater than 0.150 (the mean absorbances plus three standard deviations of 100 healthy individuals).
The kit manufactured by Orange Life in Brazil was used for immunochromatography technique (NDO-LID), adding 10μL of plasma followed by 100μL running buffer (2–3 drops). The test result was considered positive when they presented color in the test and control lines. The result was considered negative when no color was seen in the test line. The tests were photographed using a DSC-HX9V camera (Sony, Tokyo, Japan) 20min after the addition of the buffer solution.
DNA extractionThe salting out technique was used to obtain DNA from the leukocytes. The concentration of the sample was adjusted at 20ng/mL and stored at −80°C in a freezer until use.
HLA typingTyping of class I (loci A*, B*and C*) and class II (loci DRB1*, DQA1*, DQB1*) HLA alleles was conducted using the LabType™ technique (One-Lambda, USA), which applies Luminex™ technology to the reverse DNA-SSO typing method.
The principle of the technique consists in the amplification of the HLA region by PCR using a specific primer for each locus. The PCR product is biotynilated, allowing it to be detected using streptavidin conjugated with R-phycoerythrin (SAPE). The PCR product is denatured and re-hybridized in a specific sequence oligonucleotide probes immobilized in fluorescence-coded microspheres. The fluorescence intensity of PE (phycoerythrin) in each microsphere was identified by a LabScan™ 100 flow analyzer. An analysis software (HLA Fusion) was used to determine the HLA genotyping, which is based on the reaction pattern compared to the patterns associated to already identified HLA gene sequences.
Ethical aspectsThis study was approved by the Research Ethics Committee on Human Beings and by the National Research Ethics Commission (CONEP) under opinion nos. 940.163 and 1.018.673, respectively.
Statistical analysisThe frequency of alleles was obtained by direct count.
The analysis of the association between HLA alleles and NDO-LID results was conducted using the chi-square or Fisher's exact test using the Graph Pad program (http://www.graphpad.com/quickcalcs/contingency1.cfm). Statistical significance was considered for p-values less 0.05. Odds Ratio (OR) with a 95% confidence interval (95%CI) was calculated using SISA (http://www.quantitativeskills.com/sisa/) when p was statistically significant.
From the HLA genotypes for the different loci, Hardy–Weinberg equilibrium calculations were performed and the population structure was evaluated by applying the G test.
Given the variability of HLA specificities, HLA alleles were presented in allelic groups to avoid the over stratification of the sample and to perform the comparative analyses between HLA alleles, NDO-LID, and PGL-1.
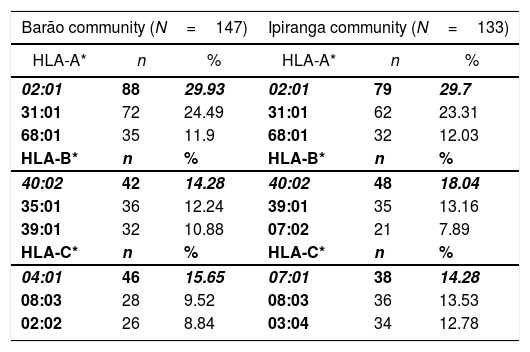
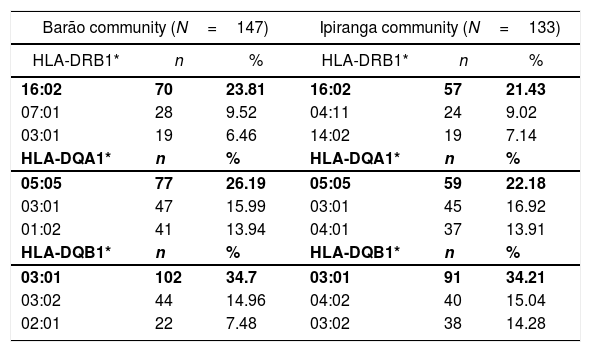
ResultsGenetic aspects of HLATyping of HLA identified 21 alleles on locus A, 52 on locus B, 22 on locus C, 42 on locus DRB1, 13 on locus DQA1, and 15 on locus DQB1. Tables 1 and 2 list the most frequent alleles in this population, and other alleles are presented in supplementary Tables (S1–S6). The most frequent class I HLA alleles in the Barão community were A*02:01 (29.93%), B*40:02 (14.28%), and C*04:01 (15.65%); and for class II: DRB1*16:02 (23.81%), DQA1*05:05 (26.19%), and DQB1*03:01 (34.7%). The same alleles were observed at Ipiranga community, namely, A*02:01 (29.7%), B*40:02 (18.04%), DRB1*16:02 (21.43%), DQA1*05:05 (22.18%) and DQB1*03:01 (34.21%); except for locus C, in which the most frequent allele was HLA-C*07:01(14.28%).
Most frequent alleles of HLA class I (loci A*, B* e C*) in Barão and Ipiranga communities.
| Barão community (N=147) | Ipiranga community (N=133) | ||||
|---|---|---|---|---|---|
| HLA-A* | n | % | HLA-A* | n | % |
| 02:01 | 88 | 29.93 | 02:01 | 79 | 29.7 |
| 31:01 | 72 | 24.49 | 31:01 | 62 | 23.31 |
| 68:01 | 35 | 11.9 | 68:01 | 32 | 12.03 |
| HLA-B* | n | % | HLA-B* | n | % |
| 40:02 | 42 | 14.28 | 40:02 | 48 | 18.04 |
| 35:01 | 36 | 12.24 | 39:01 | 35 | 13.16 |
| 39:01 | 32 | 10.88 | 07:02 | 21 | 7.89 |
| HLA-C* | n | % | HLA-C* | n | % |
| 04:01 | 46 | 15.65 | 07:01 | 38 | 14.28 |
| 08:03 | 28 | 9.52 | 08:03 | 36 | 13.53 |
| 02:02 | 26 | 8.84 | 03:04 | 34 | 12.78 |
N, number of individuals; n, number of alleles (2n); %, allele frequency; boldface, most frequent allele.
Most frequent alleles of HLA class II (DRB1*, DQA1*, DQB1*) in Barão and Ipiranga communities.
| Barão community (N=147) | Ipiranga community (N=133) | ||||
|---|---|---|---|---|---|
| HLA-DRB1* | n | % | HLA-DRB1* | n | % |
| 16:02 | 70 | 23.81 | 16:02 | 57 | 21.43 |
| 07:01 | 28 | 9.52 | 04:11 | 24 | 9.02 |
| 03:01 | 19 | 6.46 | 14:02 | 19 | 7.14 |
| HLA-DQA1* | n | % | HLA-DQA1* | n | % |
| 05:05 | 77 | 26.19 | 05:05 | 59 | 22.18 |
| 03:01 | 47 | 15.99 | 03:01 | 45 | 16.92 |
| 01:02 | 41 | 13.94 | 04:01 | 37 | 13.91 |
| HLA-DQB1* | n | % | HLA-DQB1* | n | % |
| 03:01 | 102 | 34.7 | 03:01 | 91 | 34.21 |
| 03:02 | 44 | 14.96 | 04:02 | 40 | 15.04 |
| 02:01 | 22 | 7.48 | 03:02 | 38 | 14.28 |
N, number of individuals; n, number of alleles (2n); %, allele frequency; boldface, most frequent allele.
Regarding the Hardy–Weinberg equilibrium calculations, the Barão sample showed significant values for HLA-A, HLA-B and HLA-C loci, while in the Ipiranga sample only HLA-A locus was in equilibrium.
The results of the G test showed the existence of population structure and low gene flow in the analyzed populations, and similarly the F statistic elements (frequency of alleles and presence of exclusive alleles) corroborated these findings. Due to this structure, allele frequencies were presented separately in each population.
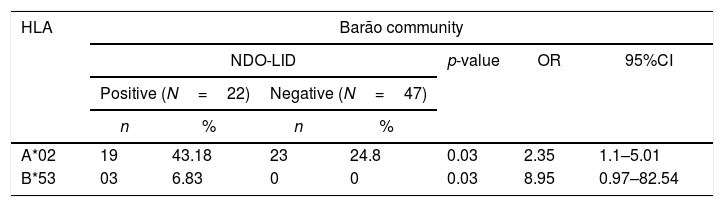
Association between NDO-LID and HLAA significant association between positive NDO-LID test was found with alleles HLA-A*02 (43.18% vs. 24.5%, p=0.03, OR=2.35) and HLA-B*53 (6.83% vs. 0.0%, p=0.03, OR=8.95) in residents of Barão community. For Ipiranga community, a significant association was found between allele HLA-B*15 (3.85% vs. 16.25%, p=0.05, OR=0.19) with negative result for the NDO-LID test. When analyzing the two populations jointly, there was an association between seropositivity to the test and the HLA-B*57 allele (6.25% vs. 1.15%, p=0.03, OR=5.73), and seronegativity with alleles HLA-B*15 (6.25% vs.14.37, p=0.005, OR=0.39) and HLA-C*02 (2.08% vs.10.92%, p=0.008, OR=0.17) (Table 3).
Significant associations between the NDO-LID test and HLA alleles of indigenous residents of Barão and Ipiranga communities (n=135), Acre state, Brazil.
| HLA | Barão community | ||||||
|---|---|---|---|---|---|---|---|
| NDO-LID | p-value | OR | 95%CI | ||||
| Positive (N=22) | Negative (N=47) | ||||||
| n | % | n | % | ||||
| A*02 | 19 | 43.18 | 23 | 24.8 | 0.03 | 2.35 | 1.1–5.01 |
| B*53 | 03 | 6.83 | 0 | 0 | 0.03 | 8.95 | 0.97–82.54 |
| HLA | Ipiranga community | ||||||
|---|---|---|---|---|---|---|---|
| NDO-LID | p-value | OR | 95%CI | ||||
| Positive (N=26) | Negative (N=40) | ||||||
| n | % | n | % | ||||
| B*15 | 2 | 3.85 | 13 | 16.25 | 0.05 | 0.19 | 0.04–0.92 |
| HLA | Barão and Ipiranga communities | ||||||
|---|---|---|---|---|---|---|---|
| NDO-LID | p-value | OR | 95%CI | ||||
| Positive (N=48) | Negative (N=87) | ||||||
| n | % | n | % | ||||
| B*15 | 6 | 6.25 | 25 | 14.37 | 0.005 | 0.39 | 0.15–1.0 |
| B*57 | 6 | 6.25 | 2 | 1.15 | 0.03 | 5.73 | 1.13–28.99 |
| C*02 | 2 | 2.08 | 19 | 10.92 | 0.008 | 0.17 | 0.04–0.76 |
NDO-LID.
HLA, human leukocyte antigen.
N, number of individuals; n, number of alleles (2n); %, allele frequency; p, Fisher's exact test (p≤0.05).
OR, odds ratio.
95%CI, 95% confidence interval.
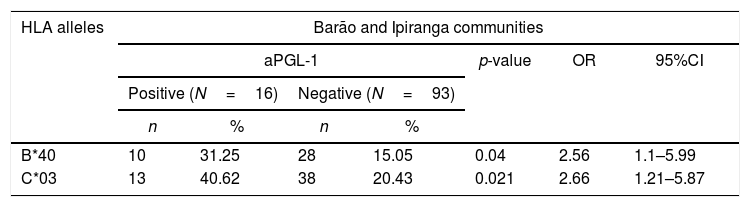
Alleles HLA-B*40 (31.25% vs. 15.15%, p=0.04, OR=2.56) and HLA-C*03 (40.62% vs. 20.43%, p=0.021, OR=2.66) were associated with a positive serological response to the ELISA PGL-1 test at Barão and Ipiranga communities (Table 4). The reduced number of participants with positive PGL-1 (16 individuals) did not enable the assessment of the association in each population separately.
A significant association between the ELISA PGL-1 test and HLA alleles on residents of Barão and Ipiranga communities, Mâncio Lima, Acre, Brazil, 2018 (n=109).
| HLA alleles | Barão and Ipiranga communities | ||||||
|---|---|---|---|---|---|---|---|
| aPGL-1 | p-value | OR | 95%CI | ||||
| Positive (N=16) | Negative (N=93) | ||||||
| n | % | n | % | ||||
| B*40 | 10 | 31.25 | 28 | 15.05 | 0.04 | 2.56 | 1.1–5.99 |
| C*03 | 13 | 40.62 | 38 | 20.43 | 0.021 | 2.66 | 1.21–5.87 |
a-PGL-1, Anti-PGL-1 ELISA.
HLA, human leukocyte antigen.
N, number of individuals; n, number of alleles (2n); %, allele frequency; p, Fisher's exact test (p≤0.05).
OR, odds ratio.
95%CI, 95% confidence interval.
The knowledge of HLA alleles is an important instrument to understand the origin of populations, and their determination contributes to the understanding of mechanisms associated with susceptibility or resistance to certain diseases.7,31,32
This study showed class I and II HLA alleles found in the population of the Puyanawa indigenous reserve. The most frequent alleles were HLA-A*02:01, HLA-B*40:02, HLA-C*04:01, HLA-C*07:01, HLA-DRB1*16:02, HLA-DQA1*05:05, and HLA-DQB1*03:01.
The finding of lack of equilibrium for the different loci in both populations was not apparently due to inbreeding processes, but, in contrast, the HLA-A locus at the Barão community and HLA-DQB1 at the Ipiranga community indicated a slight increased number of heterozygotes.
Although the structuring was observed, as well as a low gene flow among the analyzed populations (G test and F statistic), there has been no loss of allele diversity or crossbreeding that would direct the population to eventual homozygosis. Even being of the same ethnicity, marriage between individuals from these two villages is not very frequent, which explains the low gene flow observed.
According to data from Rede Brasil de Imunogenética (Brazil Immunogenetics Network), alleles HLA-A*02, HLA-B*40, and HLA-DRB1*16:02 are among the most frequent in the population of Acre. These results demonstrate the similarity between the Puyanawa indigenous population and the population of the state of Acre, showing the miscegenation between these populations.
The history of the Puyanawa people is marked by a long process of domination, enslavement, and disappearance of their culture and language. Like many peoples of Acre, they suffered from the growth of the extraction of natural rubber in the region in the early 20th century. Since the first contacts with non-indigenous peoples, many have died in conflicts or from diseases acquired in this process.33
The HLA class I and II alleles most frequently found in this study had already been described in Terena indigenous populations, who mostly inhabit the state of Mato Grosso do Sul34 except for allele HLA-DQA1*05:05, which is present among Guarani and Kaingang indigenous people31 and Cayapa people in Ecuador.35
Alleles DRB1*16:02 and DQB1*03:01 are frequent among the indigenous populations of Xavantes, Guarani Kaiowá, Guarani Mbya, Guarani Nandeva, Kaingang, and Ticuna in Brazil,31 and Mayan peoples in Guatemala.36
The Brazilian population has a great allelic diversity resulting from the miscegenation among Caucasians, Africans, and indigenous peoples.37,38 The identification of HLA alleles shows Italian immigration to the southern region of Brazil, Dutch to the Northeast, and Moroccan Jews in the Amazon region.38 During the period known as the rubber cycle, the state of Acre received many migrants from the Northeast region – mainly from Ceará – due to a drought in the late 19th century, as well as individuals from São Paulo and Syria/Lebanon.39
A study on the frequency of different HLA-DRB1 alleles in individuals from endemic areas for malaria in the North region of Brazil, including a municipality in Acre, identified that the most frequent alleles were HLA-DRB1*04, DRB1*08, DRB1*07, and DRB1*13 and that the high prevalence of allele HLA-DRB1*04 reflects the contribution from indigenous peoples to the genetic composition of the Brazilian population – especially in the north of the country40 – since this allele is characteristic of indigenous peoples of the Americas.41 These results were also observed in group evaluated in our study. Considering both populations together, the frequency of the HLA-DRB1*04 allelic group was 34.35%, with HLA-DRB1*04:11 being the most frequent. The frequencies of alleles HLA-DRB1*07, DRB1*08, and DRB1*13 were 15.91%, 17.29%, and 19.34%, respectively.
The distribution of HLA alleles and haplotypes in ethnic groups that compose the Brazilian Registry of Voluntary Bone Marrow Donors (REDOME) presents frequent alleles in indigenous peoples, such as HLA-A*03:04, A*23:02, A*29:03, A*31:28, HLA-B*35:21, and states where the allele HLA-B*40:15 was only observed in the indigenous ethnic group.42 We verified the presence of HLA-B*40 as the most frequent allele; however, the genotype found in the Puyanawa population was HLA-B*40:02.
The most frequent allele in the Puyanawa population in this study was HLA-DRB1*16:02. Alleles HLA-DRB1*15 and DRB1*16 have been associated with leprosy, especially in Indians, Japanese, and Brazilians.5,8,16 HLA-DRB1*16:01 was associated with susceptibility to BL,5 whereas HLA-DRB1*16:02 to LL.43 Although the study with the Brazilian population was conducted in a sample different from that of our study, and since leprosy cases have occurred in the indigenous reserve in the past, we can hypothesize that the identification of HLA-DRB1*16:02 could suggest that these individuals would be the most predisposed to the manifestation of leprosy. Studies with greater control must be conducted to confirm these results.
To date, there are no studies assessing the association of positive serology (NDO-LID and PGL-1) for leprosy with HLA alleles. However, there are positive associations linked to these molecules with leprosy and its clinical forms and reactions due to their importance in the presentation of antigens and triggering humoral immune response. Knowing that serological methods use techniques based on the detection of specific anti-M. leprae antibodies and that seropositivity to the tests mainly show responses triggered by humoral immunity with a higher presence of antibodies against the bacillus,18,21,22 our study showed that HLA alleles contribute to seroconversion to NDO-LID and ELISA PGL-1 tests.
Barreto et al.,44 stated in their study that the chances of developing leprosy in people seropositive to the surface protein of the M. Leprae were 2.7-fold higher than in seronegative people (p<0.01), indicating that in a follow-up of 10 seropositive individuals, there is a greater than 90% probability of detecting at least one new case in two years.
Serological studies from different regions of Brazil confirmed the anti-LID-1 and anti-NDO-LID tests as diagnostic tools for the detection of multibacillary leprosy and identification of individuals with subclinical M. leprae infection.20,26
The increase in HLA-A*02 and HLA-B*53 frequencies suggest, respectively, a two- and nine-fold higher risk of having a positive serological response to the NDO-LID test, respectively. On the other hand, individuals who presented the HLA-B*15 allele were less likely to respond to the test and, consequently, have lower chances of developing MB, or even getting sick. In a distinct population, Santana et al.,6 observed the presence of HLA-B*53 (p=0.05 and pc>0.05) in patients with BL, and association of the HLA-B*15 allele (p=0.05 and pc>0.05) with the manifestation of the type 1 reaction. In the joint analysis of the two populations, there was association with HLA-B*57 increasing the chance by almost six-fold of having positive serological results to NDO-LID, whereas the HLA-B*15 and HLA-C*02 alleles would reduce this possibility.
Seronegativity does not rule out the possibility of disease development. The chance is much higher in seropositive than in seronegative individuals.21,44 According to a follow-up study in schoolchildren, 22.3% and 9.4% of seropositive and seronegative individuals, respectively, developed leprosy.44
The analysis of HLA molecules of the population of the two communities with ELISA PGL-1 showed that HLA-B*40, and HLA-C*03 alleles were associated with an almost 3-fold greater chance of presenting positive serological response. In a review published by Jarduli et al.,8 HLA-B*40 antigen and HLA-A*02-B*40, HLA-A*11-B*40 and HLA-A*24-B*40 haplotypes were frequent in Indians with leprosy.
Studies have shown that seropositivity to ELISA anti-PGL-1 associated with the risk of developing leprosy, especially in intra-domiciliary contacts of MB cases18,19,21,45 is directly related to genetic susceptibility, frequent exposure to the bacillus, and leprosy endemicity.25,45,46 Moreover, evidence also shows that increased age, health problems, and socioeconomic conditions are associated with increased disease risk.47
Thus, establishing the HLA frequency of this indigenous community was crucial to characterize the genetic profile of this population, contributing to the distribution of HLA alleles in the Brazilian population and establishing the HLA association with serological tests (NDO-LID and PGL-1) used in the diagnosis and monitoring of leprosy. The association of HLA alleles and seropositivity to surface proteins of M. leprae may be a genetic indication for the predisposition to leprosy. However, more studies are needed to further establish the role of HLA alleles in seroconversion to the rapid NDO-LID and ELISA PGL-1 tests, and so appropriately targeted public health policies are established.
FundingThis research received a specific subsidy from the following funding agencies: Fundação Paulista contra Hanseníase and Fundação de Amparo à Pesquisa do estado do Acre/Programa Pesquisa para o SUS (PPSUS). The entire resource was used to carry out data collection and statistical analysis.
Conflicts of interestThe authors declare no conflicts of interest.
We would like to thank the students and professors of the undergraduate course in Nursing of Federal University of Acre (UFAC) who participated in data collection and the group of the Laboratory of Immunology and Microbiology of UFAC for their support in the preparation of the material for analysis. To the research group of the Laboratory of Immunology and Immunogenetics of the Instituto Lauro de Souza Lima for their support and assistance in laboratory analyses and supply of antigens. To the Programa de Pesquisa para o SUS (PPSUS) of the state of Acre, and the Fundação Paulista contra Hanseníase for the financial support.