Multi-drug resistant Gram-negative bacilli (GNB) have been reported as cause of serious hospital-acquired infections worldwide. The aim of this study was to investigate the in vitro activity of ceftolozane-tazobactam compared to other agents against GNB isolated from patients admitted to Brazilian medical centers between the years 2016 and 2017. Presence of β-lactamase encoding genes was also evaluated.
MethodsAntimicrobial susceptibility testing of GNB isolated from intra-abdominal (IAI), respiratory (RTI), and urinary tract infections (UTI) was performed according to ISO 227-1 guidelines and interpreted following CLSI and BrCAST/EUCAST guidelines. Qualifying Enterobacteriaceae isolates were screened for the presence of β-lactamase genes by PCR followed by DNA sequencing.
Results1748 GNB collected from UTI (45.2%), IAI (25.7%) and RTI (29.1%) were evaluated. Ceftolozane-tazobactam remained highly active (94.7%) against E. coli isolates. Among K. pneumoniae, susceptibility rates were 85.9% and 85.4% for amikacin and colistin, whereas ceftolozane-tazobactam (44.1% susceptible) and carbapenems (55.2-62.2% susceptible) showed poor activity due to blaKPC-2. Against E. cloacae amikacin, imipenem, and meropenem retained good activity (>90%). Ceftolozane-tazobactam was the most potent β-lactam agent tested against P. aeruginosa (90.9% susceptible), including ceftazidime and imipenem resistant isolates. β-lactamase encoding genes testing was carried out in 433 isolates. blaCTX-M variants were predominant in E. coli, P. mirabilis and E. cloacae. Among the K. pneumoniae molecularly tested, most carried blaKPC (68.5%), with all harboring blaKPC-2, except two isolates carrying blaKPC-3 or blaKPC-30. ESBL encoding genes, mainly CTX-M family, were frequently detected in K. pneumoniae, plasmid-mediated AmpC were rare. A variety of PDC encoding genes were detected in P. aeruginosa isolates with five isolates harboring MBL and one KPC encoding genes.
ConclusionCeftolozane-tazobactam was very active against E. coli, P. mirabilis and P. aeruginosa isolates and could constitute an excellent therapeutic option including for those isolates resistant to extended-spectrum cephalosporins and carbapenems but not producers of carbapenemases.
In 2019, the World Health Organization has identified antimicrobial resistance as one of the world’s top 10 global health threats1https://www.who.int/emergencies/ten-threats-to-global-health-in-2019. The U.S. Centers for Disease Control and Prevention (CDC) has recently reported that more than 2.8 million antibiotic-resistant infections occur in the United States each year.2 Although antimicrobial resistance varies widely depending on the bacterial species, antimicrobial agent and geographical region, high levels of antimicrobial resistance for several bacterial species–antimicrobial combinations have been also observed among invasive isolates reported to the European Antimicrobial Resistance Surveillance Network (EARS-Net).3 Unfortunately, Gram-negative bacilli (GNB) exhibiting resistance to multiple antimicrobial agents have been reported as important causes of serious hospital acquired infections in Brazilian hospitals.4 Carbapenems used to be considered drugs of choice for treatment of multi-drug resistant GNB5; however, the emergence and spread of carbapenemase encoding genes carried by mobile genetic elements has jeopardized the clinical usefulness of this important therapeutic class of antimicrobials.5 The last report of the Brazilian Health Surveillance Agency estimated that 44.1% of Klebsiella pneumoniae and 42.9% of Pseudomonas aeruginosa isolates causing catheter-related bloodstream infections in ICU adult patients were resistant to carbapenems.4 Although many Brazilian medical centers have been aware of their carbapenem resistance rates, the underlying mechanisms of carbapenem resistance are largely unknown.
The Study for Monitoring Antimicrobial Resistance Trends (SMART) program has generated data on the frequency of antimicrobial susceptibility of GNB associated with urinary tract (UTI), intra-abdominal (IAI) and respiratory tract (RTI) infections worldwide since 2008. The principal aim of this study was to determine the frequency of pathogens and in vitro activity of ceftolozane-tazobactam compared to other agents against the most frequently identified GNB isolated from patients admitted to Brazilian medical centers between the years 2016 and 2017. The presence of β-lactamase encoding genes, the main mechanism of β-lactam resistance, was also determined for selected isolates.
MethodsBacterial isolatesNon-duplicate GNB isolates were collected consecutively from eight Brazilian medical centers between January 1, 2016, and December 31, 2017. The participating medical centers were located in the Brazilian cities of Rio de Janeiro (one center), Salvador (one center), and São Paulo (six centers). Each participating medical center collected up to 100 consecutive GNB isolated from patients with intra-abdominal (IAI) and respiratory (RTI) infections, and 50 GNB from urinary-tract infections (UTI) per year. GNB were identified at the species level at the respective participant medical center and shipped to a central microbiology laboratory (International Health Management Associates, IHMA, Schaumburg, IL, USA), where confirmation of bacterial species, antimicrobial susceptibility testing, and molecular characterization of β-lactamase encoding genes were carried out. Bacterial identification at the species level was confirmed for all isolates using MALDI-TOF spectrometry (Bruker Daltonics, Billerica, MA, USA).
Susceptibility testingAntimicrobial susceptibility testing for amikacin, aztreonam, cefepime, cefotaxime, ceftazidime, ceftolozane-tazobactam, ceftriaxone, ciprofloxacin, colistin, ertapenem, imipenem, meropenem, and piperacillin-tazobactam was determined by testing customized MicroScan dehydrated broth microdilution panels (Siemens Medical Solutions Diagnostics, West Sacramento, CA, USA) according to the ISO 227-1 guidelines and interpreted following both CLSI7 and BrCAST/EUCAST8,9 guidelines. Quality control of broth microdilution panels followed the manufacturer’s and CLSI guidelines using the following ATCC strains: E. coli ATCC 25922, P. aeruginosa ATCC 27853, and K. pneumoniae ATCC 700603. Corresponding QC values tested were within the acceptable ranges as specified by CLSI. E. coli and Klebsiella pneumoniae isolates with MICs ≥2μg/mL for ceftazidime, ceftriaxone, or aztreonam were screened as “ESBL phenotype”. Enterobacteriaceae with MIC≥4μg/mL for imipenem and/or meropenem were defined as carbapenem resistant. P. aeruginosa isolates having MICs >8μg/mL and >2μg/mL, respectively, were classified as not susceptible (NS) to ceftazidime and meropenem.
Molecular characterization of β-lactamase encoding genesAll Enterobacteriaceae with MICs ≥4mg/L for ceftolozane-tazobactam and/or ≥1mg/L for ertapenem (except Proteeae Enterobacteriaceae) and/or ≥2mg/L for imipenem were selected for characterization of β-lactamase content as well as P. aeruginosa isolates displaying ceftolozane-tazobactam MICs ≥8mg/L and/or imipenem MICs ≥4mg/L. For comparison reasons, 50% of the E. coli and K. pneumoniae possessing the ESBL phenotype but showing susceptibility to ertapenem, imipenem, and ceftolozane-tazobactam were also selected for molecular characterization of β-lactamase encoding genes. Qualifying Enterobacteriaceae isolates were screened for the presence of β-lactamase genes (bla) encoding extended spectrum-β-lactamases (ESBLs; TEM, SHV, CTX-M, VEB, PER, GES), AmpC β-lactamases (ACC, ACT, CMY, DHA, FOX, MIR, MOX), and carbapenemases (KPC, OXA-48-like, IMP, VIM, NDM, SPM, and GIM) by multiplex PCR as described previously.8 Limited sequencing was performed on blaTEM and blaSHV to identify genes encoding TEM-type and SHV-type enzymes containing amino acid substitutions common to ESBLs (SHV A146V, G238S, G238A, E240K; TEM E104K, R164S, R164C, R164H, G238S). All detected genes, excluding blaSHV and blaTEM that did not encode ESBLs and the intrinsic, chromosomally coded blaAmpC of Citrobacter spp. (blaCMY-type) and Enterobacter spp. (blaACT-type and blaMIR-type), were amplified and sequenced in their entirety. Qualifying P. aeruginosa isolates were screened for the presence of bla encoding ESBLs (TEM, SHV, CTX-M, VEB, PER, GES), plasmid-encoded AmpC β-lactamases (ACC, ACT, CMY, DHA, FOX, MIR, MOX), and carbapenemases (KPC, OXA-24-like, IMP, VIM, NDM, SPM, and GIM) by multiplex PCR as described. All detected genes, as well as the chromosomally encoded Pseudomonas-derived cephalosporinase blaAmpC (PDC) common to the species, were amplified and sequenced in their entirety.
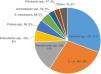
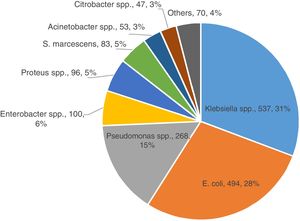
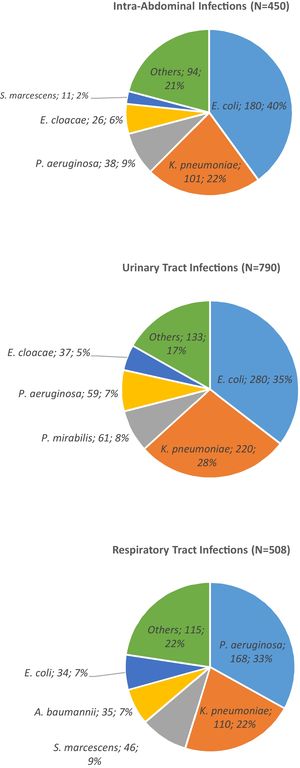
ResultsA total of 1,748 GNB were collected at the eight Brazilian medical centers between the years 2016 (N=776) and 2017 (N=972). Most isolates were recovered from female patients (51%) of all ages. Approximately 77.9% (1362 of 1748) of GNB belonged to five bacterial species as depicted in Fig. 1. The distribution of the five most relevant bacterial species according to the body site of infection is shown in Fig. 2. Most of these pathogens were collected from urinary tract (45.2%), followed by respiratory (29.1%), and intra-abdominal infections (25.7%). In general, among all species isolated, E. coli (n=494) and K. pneumoniae (n=431) were the most frequently identified species from all sources of infection. While E. cloacae, E. coli, K. pneumoniae, and P. mirabilis were more frequently isolated from urine tract infections, P. aeruginosa (33.0%) was the most frequent species recovered from respiratory tract infections, followed by K. pneumoniae (23.0%), S. marcescens (9%), and A. baumannii (7%). E. coli (40.0%) was the most frequent species isolated from patients with intra-abdominal infections followed by K. pneumoniae (22.0%) as depicted in Fig. 2.
Distribution of isolates according to the bacterial species collected from participating Brazilian medical centers of the SMART Program (Brazil, 2016-2017).
a. Acinetobacter spp. (53): A. baumannii (49), A. ursingii (1), A. guillouiae (1), A. nosocomialis (1), and A. pittii (1);
b. Citrobacter spp. (47): C. amalonaticus (1), C. farmeri (3), C. freundii (30), and C. koseri (13);
c. Enterobacter spp. (100): E. asburiae (13), E. cloacae (81), E. kobei (5), and Enterobacter spp. (1);
d. Klebsiella spp. (537): K. aerogenes (65), K. oxytoca (19), K. pneumoniae (431), and K. variicola (22);
e. Proteus spp. (96): P. hauseri (2), P. mirabilis (91), and P. vulgaris (3);
f. Pseudomonas spp. (268): P. aeruginosa (265), P. mosselii (1), P. otitidis (1), and P. putida (1);
g. Others (70): Achromobacter xylosoxidans (5), Aeromonas caviae (1), Aeromonas hydrophila (1), Burkholderia cenocepacia (3), Escherichia hermanii (1), Morganella morganii (20), Pluralibacter gergoviae (2), Providencia alcalifaciens (1), Providencoa rettgeri (4), Providencia stuartii (6), Raoultella ornithinolytica (1), and Raoutella planticola (1), Salmonella spp. (2), Stenotrophomonas maltophilia (22).
Antimicrobial susceptibility profiles of the five most relevant pathogens causing infections in the participating Brazilian medical centers is shown in Table 1. For comparison purposes the susceptibility and resistance rates according to the CLSI and EUCAST breakpoints are shown in Table 1; however, since BrCAST/EUCAST criteria have been recommended by the Brazilian Ministry of Health,10 the BrCAST/EUCAST susceptibility/resistance rates were considered when evaluating the antimicrobial activity of the respective antimicrobial agents, when these percentages varied.
Antimicrobial susceptibility profile of the five most frequent pathogens causing infections at participating Brazilian medical centers of the SMART Program (Brazil, 2016-2017).
| PathogenAntimicrobial Agents | Broth Microdiluition(μg/mL) | CLSIa | EUCASTa | |||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | S (%) | R (%) | S (%) | R (%) | |
| Enterobacter cloacae (81) | ||||||
| Amikacin | ≤ 4 | ≤ 4 | 98.8 | 1.2 | 98.8 | 1.2 |
| Aztreonam | > 16 | > 16 | 39.5 | 60.5 | 38.3 | 60.7 |
| Cefepime | 4 | > 32 | 46.9 | 53.1 | 39.5 | 60.5 |
| Ceftazidime | 32 | > 32 | 39.5 | 60.5 | 35.8 | 61.2 |
| Ceftolozane-tazobactam | 2 | 16 | 59.3 | 40.7 | 46.9 | 53.1 |
| Ceftriaxone | > 32 | > 32 | 38.3 | 61.7 | 38.3 | 60.7 |
| Ciprofloxacin | ≤ 0.25 | > 2 | 61.7 | 38.3 | 61.7 | 38.3 |
| Colistin | ≤ 1 | > 1 | 95.1 | 4.9 | 95.1 | 4.9 |
| Ertapenem | ≤ 0.12 | 1 | 77.8 | 22.2 | 77.8 | 22.2 |
| Imipenem | ≤ 0.5 | 1 | 96.3 | 3.7 | 98.8 | 1.2 |
| Meropenem | ≤ 0.12 | 0.25 | 98.8 | 1.2 | 98.8 | 1.2 |
| Piperacillin-tazobactam | 16 | > 64 | 50.6 | 49.4 | 43.2 | 56.8 |
| E. coli (494) | ||||||
| Amikacin | ≤ 4 | 8 | 98.6 | 0.4 | 97.4 | 2.6 |
| Aztreonam | ≤ 1 | > 16 | 80.0 | 20.0 | 71.5 | 28.5 |
| Cefepime | ≤ 1 | > 32 | 76.5 | 23.5 | 73.3 | 26.7 |
| Ceftazidime | ≤ 1 | 16 | 84.4 | 15.6 | 78.1 | 21.9 |
| Ceftolozane-tazobactam | 0.25 | 0.5 | 96.8 | 3.2 | 94.7 | 5.3 |
| Ceftriaxone | ≤ 1 | > 32 | 71.7 | 28.3 | 71.7 | 28.3 |
| Ciprofloxacin | ≤ 0.25 | > 2 | 55.1 | 44.9 | 55.1 | 44.9 |
| Colistin | ≤ 1 | ≤ 1 | 99.4 | 0.6 | 99.4 | 0.6 |
| Ertapenem | ≤ 0.06 | ≤ 0.06 | 97.8 | 2.2 | 97.8 | 2.2 |
| Imipenem | ≤ 0.5 | ≤ 0.5 | 98.6 | 0.6 | 99.4 | 0.6 |
| Meropenem | ≤ 0.12 | ≤ 0.12 | 99.0 | 1.0 | 99.0 | 1.0 |
| Piperacillin-tazobactam | ≤ 2 | 8 | 93.3 | 6,7 | 90.9 | 9.1 |
| K. pneumoniae (431) | ||||||
| Amikacin | ≤ 4 | 32 | 89.6 | 10.4 | 85.9 | 14.1 |
| Aztreonam | > 16 | > 16 | 37.4 | 62.6 | 35.0 | 65.0 |
| Cefepime | 32 | > 32 | 36.7 | 63.3 | 35.7 | 64.3 |
| Ceftazidime | 32 | > 32 | 39.0 | 61.0 | 34.3 | 65.7 |
| Ceftolozane-tazobactam | 4 | > 32 | 49.7 | 50.3 | 44.1 | 55.9 |
| Ceftriaxone | > 32 | > 32 | 36.2 | 63.8 | 36.2 | 63.8 |
| Ciprofloxacin | > 2 | > 2 | 31.6 | 68.4 | 31.6 | 68.4 |
| Colistin | ≤ 1 | > 4 | 85.4 | 14.6 | 85.4 | 14.6 |
| Ertapenem | 0.25 | > 4 | 55.2 | 44.8 | 55.2 | 44.8 |
| Imipenem | ≤ 0.5 | > 32 | 61.0 | 39.0 | 62.2 | 37.8 |
| Meropenem | ≤ 0.12 | > 16 | 58.7 | 42.3 | 60.1 | 39.9 |
| Piperacillin-tazobactam | > 64 | > 64 | 41.5 | 58.5 | 36.7 | 63.3 |
| P. aeruginosa (265) | ||||||
| Amikacin | ≤ 4 | 16 | 91.7 | 8.3 | 87.9 | 12.1 |
| Aztreonam | 8 | > 16 | 52.5 | 47.5 | 67.6 | 32.4 |
| Cefepime | 8 | 32 | 68.3 | 31.7 | 68.3 | 31.7 |
| Ceftazidime | 4 | > 32 | 66.8 | 33.2 | 66.8 | 33.2 |
| Ceftolozane-tazobactam | 1 | 4 | 90.9 | 9.1 | 90.9 | 9.1 |
| Ciprofloxacin | ≤ 0.25 | > 2 | 69.8 | 30.2 | 69.8 | 30.2 |
| Colistin | ≤1 | 2 | 98.9 | 1.1 | 98.9 | 1.1 |
| Imipenem | 1 | 16 | 63.8 | 36.2 | 70.2 | 29.8 |
| Meropenem | 1 | > 16 | 66.0 | 34.0 | 66.0 | 34.0 |
| Piperacillin Tazobactam | 16 | > 64 | 59.6 | 40.4 | 59.6 | 40.4 |
| P. mirabilis (91) | ||||||
| Amikacin | ≤ 4 | 8 | 97.8 | 2.2 | 95.6 | 4.4 |
| Aztreonam | ≤ 1 | 2 | 94.5 | 5.5 | 83.5 | 16.5 |
| Cefepime | ≤ 1 | 32 | 82.4 | 17.6 | 80.2 | 19.8 |
| Ceftazidime | ≤ 1 | 2 | 95.6 | 4.4 | 85.7 | 14.3 |
| Ceftolozane-tazobactam | 0.5 | 0.5 | 98.9 | 1.1 | 97.8 | 2.2 |
| Ceftriaxone | ≤ 1 | > 32 | 78.0 | 22.0 | 78.0 | 22.0 |
| Ciprofloxacin | ≤ 0.25 | > 2 | 72.5 | 27.5 | 72.5 | 27.5 |
| Ertapenem | ≤ 0.06 | ≤ 0.06 | 95.6 | 4.4 | 95.6 | 4.4 |
| Imipenem | 1 | 2 | 55.0 | 45.0 | 92.3 | 7.7 |
| Meropenem | ≤ 0.12 | ≤ 0.12 | 98.9 | 1.1 | 98.9 | 1.1 |
| Piperacillin-tazobactam | ≤ 2 | ≤2 | 97.8 | 2.2 | 96.7 | 3.3 |
Colistin (MIC50/90, ≤1/≤1μg/mL; 99.4% susceptible) and imipenem (MIC50/90, ≤0.5/≤0.5μg/mL; 99.4% susceptible) were the most in vitro active agents tested against the 494 E. coli isolates, followed by meropenem (MIC50/90, ≤0.12/≤0.12μg/mL; 99.0% susceptible), ertapenem (MIC50/90, ≤0.06/≤0.06μg/mL; 97.8% susceptible), and amikacin (MIC50/90, ≤4/8μg/mL; 97.4% susceptible). In contrast, the lowest susceptibility rates were observed for ciprofloxacin (MIC50/90, ≤0.25/>2μg/mL; 55.1% susceptible), followed by aztreonam (MIC50/90, ≤1/>16μg/mL; 71.5% susceptible), ceftriaxone (MIC50/90, ≤1/> 32μg/mL; 71.7% susceptible), cefepime (MIC50/90, ≤1/>32μg/mL, 73.3% susceptible), and ceftazidime (MIC50/90, ≤1/>16μg/mL, 78.1% susceptible). Ceftolozane-tazobactam (MIC50/90, 0.25/0.5μg/mL) remained highly active against E. coli isolates, 94.7% of which were susceptible to this agent, including 87.2% of 102 E. coli isolates exhibiting the ESBL phenotype as shown in Table 2.
Susceptibility rates to distinct antimicrobial agents of the five most frequent pathogens according to the phenotype of resistance.
| E. cloacae non susceptible to ceftazidime (52) | Broth Microdiluitionμg/mL | CLSIa | EUCASTa | |||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | S (%) | R (%) | S (%) | R (%) | |
| Amikacin | ≤ 4 | ≤ 4 | 97.9 | 2.1 | 98.0 | 2.0 |
| Aztreonam | > 16 | > 16 | 0 | 100 | 3.8 | 96.2 |
| Cefepime | 16 | > 32 | 12.2 | 87.8 | 5.7 | 94;3 |
| Ceftazidime | > 32 | > 32 | 0 | 100.0 | 0 | 100.0 |
| Ceftolozane-tazobactam | 8 | 32 | 32.6 | 67.4 | 19.2 | 80.8 |
| Ceftriaxone | > 32 | > 32 | 0 | 100 | 5.7 | 94.3 |
| Ciprofloxacin | 2 | > 2 | 42.8 | 57.2 | 44.2 | 55.8 |
| Colistin | ≤ 1 | ≤ 1 | 94.2 | 5.8 | 94.2 | 5.8 |
| Ertapenem | 0.5 | 2 | 63.2 | 37.8 | 65.3 | 34,7 |
| Imipenem | ≤ 0.5 | 1 | 95.9 | 4.1 | 98.0 | 2.0 |
| Meropenem | ≤ 0.12 | 0.25 | 97.9 | 2.1 | 98.0 | 2.0 |
| Piperacillin-tazobactam | 64 | > 64 | 20.4 | 79.6 | 11.5 | 88.5 |
| ESBL-producing E. coli (102) | ||||||
| Amikacin | ≤ 4 | 8 | 97.1 | 2.9 | 94.1 | 2.9 |
| Aztreonam | > 16 | > 16 | 22.6 | 77.4 | 2.9 | 97.1 |
| Cefepime | 32 | > 32 | 5.8 | 94.2 | 0 | 100.0 |
| Ceftazidime | 8 | 32 | 37.3 | 62.7 | 21.5 | 78.5 |
| Ceftolozane-tazobactam | 0.5 | 2 | 92.2 | 7.8 | 87.2 | 12.8 |
| Ceftriaxone | > 32 | > 32 | 0 | 100.0 | 0 | 100.0 |
| Ciprofloxacin | > 2 | > 2 | 19.6 | 80.4 | 19.6 | 80.4 |
| Colistin | ≤ 1 | ≤ 1 | 99.0 | 1.00 | 99.0 | 1.00 |
| Ertapenem | ≤ 0.06 | 0.12 | 97.1 | 2.9 | 97.0 | 3.0 |
| Imipenem | ≤ 0.5 | ≤ 0.5 | 99.0 | 1.0 | 99.0 | 1.0 |
| Meropenem | ≤ 0.12 | ≤ 0.12 | 99.0 | 1.0 | 99.0 | 1.0 |
| Piperacillin-tazobactam | 4 | 32 | 86.3 | 13.7 | 80.3 | 19.7 |
| ESBL-producing K. pneumoniae (144) | ||||||
| Amikacin | ≤ 4 | 16 | 92.4 | 7.6 | 88,9 | 11.1 |
| Aztreonam | > 16 | > 16 | 2.1 | 97.9 | 0.7 | 99.3 |
| Cefepime | > 32 | > 32 | 0.7 | 99.3 | 0.7 | 99.3 |
| Ceftazidime | > 32 | > 32 | 4.2 | 95.8 | 0.7 | 99.3 |
| Ceftolozane-tazobactam | 16 | > 32 | 36.1 | 63.9 | 22.9 | 77.1 |
| Ceftriaxone | > 32 | > 32 | 0.7 | 99.3 | 0.7 | 99.3 |
| Ciprofloxacin | > 2 | > 2 | 2.1 | 97.9 | 2.1 | 97.9 |
| Colistin | ≤ 1 | > 4 | 79.9 | 20.1 | 79.9 | 20.1 |
| Ertapenem | 0.5 | > 4 | 53.5 | 46.5 | 53.5 | 46.5 |
| Imipenem | ≤ 0.5 | > 32 | 63.2 | 36.8 | 66.0 | 44.0 |
| Meropenem | ≤ 0.12 | > 16 | 60.4 | 39.6 | 63.2 | 36.8 |
| Piperacillin-tazobactam | > 64 | > 64 | 22.2 | 77.8 | 13.2 | 86.8 |
| K. pneumoniae non susceptible to imipenem (168) | ||||||
| Amikacin | ≤ 4 | > 32 | 79.2 | 20.8 | 69.9 | 30.1 |
| Aztreonam | > 16 | > 16 | 1.2 | 98.8 | 0.0 | 100.0 |
| Cefepime | > 32 | > 32 | 0.6 | 99.4 | 0.0 | 100.0 |
| Ceftazidime | > 32 | > 32 | 2.4 | 97.6 | 0.6 | 99.4 |
| Ceftolozane-tazobactam | > 32 | > 32 | 1.2 | 98.8 | 0.6 | 99.4 |
| Ceftriaxone | > 32 | > 32 | 0.0 | 100.0 | 0.0 | 100.0 |
| Ciprofloxacin | > 2 | > 2 | 3.0 | 97.0 | 2.5 | 97.5 |
| Colistin | ≤ 1 | > 4 | 71.4 | 28.6 | 71.4 | 28.6 |
| Ertapenem | > 4 | > 4 | 1.2 | 98.8 | 0.6 | 99.4 |
| Imipenem | 32 | > 32 | 0.0 | 100.0 | 0.0 | 100.0 |
| Meropenem | > 16 | > 16 | 1.8 | 98.2 | 0.6 | 99.4 |
| Piperacillin-tazobactam | > 64 | > 64 | 1.2 | 98.8 | 0.6 | 99.4 |
| P. aeruginosa non susceptible to ceftazidime (88) | ||||||
| Amikacin | ≤ 4 | > 32 | 85.2 | 14.8 | 77.3 | 22.7 |
| Aztreonam | > 16 | > 16 | 11.4 | 88.6 | 29.6 | 70.4 |
| Cefepime | 32 | > 32 | 19.3 | 80.7 | 19.3 | 80.7 |
| Ceftazidime | > 32 | > 32 | 0.0 | 100.0 | 0.0 | 100.0 |
| Ceftolozane-tazobactam | 4 | 32 | 73.9 | 26.1 | 73.9 | 26.1 |
| Ciprofloxacin | 1 | > 2 | 47.7 | 52.3 | 47.7 | 52.3 |
| Colistin | ≤ 1 | 2 | 96.6 | 3.4 | 96.6 | 3.4 |
| Imipenem | 2 | 32 | 50.0 | 50.0 | 53.4 | 46.6 |
| Meropenem | 4 | > 16 | 44.3 | 55.7 | 44.3 | 55.7 |
| Piperacillin-tazobactam | > 64 | > 64 | 8.0 | 92.0 | 8.0 | 92.0 |
| P. aeruginosa non susceptible to imipenem (96) | ||||||
| Amikacin | ≤ 4 | > 32 | 86.5 | 13.5 | 77.2 | 22.8 |
| Aztreonam | 16 | > 16 | 30.2 | 69.8 | 46.8 | 53.2 |
| Cefepime | 8 | > 32 | 50.0 | 50.0 | 43.0 | 57.0 |
| Ceftazidime | 8 | > 32 | 54.2 | 45.8 | 48.1 | 51.9 |
| Ceftolozane-tazobactam | 2 | 16 | 82.3 | 17.7 | 79.8 | 20,2 |
| Ciprofloxacin | 0.5 | > 2 | 53.1 | 46.9 | 48.1 | 51.9 |
| Colistin | ≤ 1 | 2 | 99.0 | 1.0 | 99.0 | 1.0 |
| Imipenem | 8 | 32 | 0.0 | 82.3 | 0.0 | 100,0 |
| Meropenem | 16 | > 16 | 13.5 | 86.5 | 3.8 | 96.2 |
| Piperacillin-tazobactam | 32 | > 64 | 41.7 | 58.3 | 32.9 | 67.1 |
| P. mirabilis ESBL (10) | ||||||
| Amikacin | ≤ 4 | 8 | 100.0 | 0.0 | 100.0 | 0.0 |
| Aztreonam | 2 | 16 | 70.0 | 30.0 | 30.0 | 30.0 |
| Cefepime | 32 | > 32 | 0.0 | 100.0 | 0,0 | 100.0 |
| Ceftazidime | 2 | 8 | 80.0 | 20.0 | 40.0 | 60,0 |
| Ceftolozane-tazobactam | 0.5 | 1 | 100.0 | 0.0 | 90.0 | 10.0 |
| Ceftriaxone | > 32 | > 32 | 0.0 | 100.0 | 0.0 | 100,0 |
| Ciprofloxacin | > 2 | > 2 | 0.0 | 100.0 | 0.0 | 100.0 |
| Ertapenem | ≤ 0.06 | ≤ 0.06 | 100.0 | 0.0 | 100.0 | 0,0 |
| Imipenem | 1 | 2 | 60.0 | 40.0 | 100.0 | 0.0 |
| Meropenem | ≤ 0.12 | ≤ 0.12 | 100.0 | 0.0 | 100.0 | 0.0 |
| Piperacillin-tazobactam | ≤ 2 | 8 | 100.0 | 0.0 | 90.0 | 0.0 |
Among the 431 K. pneumoniae evaluated in this study, the highest susceptibility rates were observed for amikacin (MIC50/90, ≤4/32μg/mL, 85.9% susceptible) and colistin (MIC50/90, ≤1/>4μg/mL, 85.4% susceptible). Ceftolozane-tazobactam (MIC50/90, ≤4/32μg/mL, 44.1% susceptible) as well as carbapenems (55.2-62.2% susceptible) showed poor activity against isolates of K. pneumoniae as displayed in Table 1. While 22.9% of the 144 K. pneumoniae exhibiting the ESBL phenotype remained susceptible to ceftolozane-tazobactam, 98.8% of the 168 K. pneumoniae non-susceptible to imipenem were resistant to this combination, as shown in Table 2. In fact, against these pathogens, only amikacin and colistin showed susceptibility rates superior to 70%.
A total of 81 E. cloacae were analyzed in this study. Among the extended cephalosporins, ceftolozane-tazobactam (MIC50/90, 2/16μg/mL, 46.9% susceptible) showed the highest susceptibility rate against this species followed by cefepime (MIC50/90, 4/>32μg/mL, 39.5% susceptible), ceftriaxone (MIC50/90, >32/>32μg/mL, 38.3% susceptible), and ceftazidime (MIC50/90, 32/>32μg/mL, 35.8% susceptible). In contrast, imipenem (MIC50/90, ≤0.5/1μg/mL, 98.8% susceptible), meropenem (MIC50/90, ≤0.12/0.25μg/mL, 98.8% susceptible), and amikacin (MIC50/90, ≤4/≤4μg/mL, 98.8% susceptible) were active against most E. cloacae (Table 1). Colistin (MIC50/90, ≤1/>2μg/mL, 95.1% susceptible) showed good in vitro activity against E. cloacae. Against the 52 E. cloacae isolates not susceptible to ceftazidime, only amikacin, imipenem, and meropenem retained good activity against such isolates with identical susceptibility rates (98.0%) followed by colistin (94.2% susceptible) as demonstrated in Table 2.
Ceftolozane-tazobactam (MIC50/90, 0.5/0.5μg/mL; 97.8% susceptible) and meropenem (MIC50/90, ≤0.12/≤0.12μg/mL; 98.9% susceptible) were the most active antimicrobials against the 91 P. mirabilis evaluated followed by piperacillin-tazobactam (MIC50/90, ≤2/≤2μg/mL; 96.7% susceptible), ertapenem (MIC50/90, ≤0.06/≤0.06μg/mL; 95.6% susceptible), and amikacin (MIC50/90, ≤4/8μg/mL; 95.6% susceptible). Although cefepime, ceftazidime, and ceftriaxone showed good in vitro activity with MIC50s ≤1μg/mL, 19.8%, 14.3%, and 22.0% of the isolates tested were resistant to these agents, respectively (Table 1). Ertapenem, imipenem, and meropenem were highly active against the 10 P. mirabilis isolates that exhibited the ESBL phenotype (Table 2). Ceftolozane-tazobactam (MIC50, 0.5μg/mL) and piperacillin-tazobactam (MIC50, ≤2μg/mL) also inhibited nine of these isolates (Table 2).
Ceftolozane-tazobactam was the most potent (MIC50/90, 1/4μg /mL) β-lactam agent tested against 265 P. aeruginosa, inhibiting 90.9% of isolates, while ceftazidime (MIC50/90, 4/>32μg/mL) imipenem (MIC50/90, 1/16μg /mL) and meropenem (MIC50/90, 1/>16μg/mL) inhibited only 66.8%, 70.2%, and 66.0% of these isolates, respectively, as shown in Table 1. Colistin (MIC50/90, ≤1/2μg/mL; 98.9% susceptible) and amikacin (MIC50/90, ≤4/16μg/mL; 87.9% susceptible) were active against P. aeruginosa. Ceftolozane-tazobactam retained moderate activity against P. aeruginosa isolates that were non susceptible to ceftazidime or imipenem exhibiting susceptibility rates of 73.9% and 79.8%, respectively. In contrast, colistin was highly active against isolates displaying both phenotypes, ceftazidime non-susceptible (MIC50/90, ≤1/2μg/mL; 96.6% susceptible) or imipenem non-susceptible (MIC50/90, ≤1/2μg/mL; 98.7%) as shown in Table 2.
Detection of β-lactamase encoding genesAccording to the resistance profile, the molecular characterization of β-lactamase encoding genes was carried out in 433 isolates; i.e., in 36 of 81 E. cloacae (44.4%), 52 of 494 (10.5%) E. coli, 238 of 431 (55.2%) K. pneumoniae, 3 of 91 (3.3%) P. mirabilis, and 104 of 265 (39.2%) P. aeruginosa that fulfilled the study criteria specified in the material and methods section. The distribution of β-lactamase encoding genes according to bacterial species and medical center location is shown in Table 3. ESBL encoding genes were found in 37 of 52 selected E. coli. These isolates harbored a single blaCTX-M variant, with predominance of blaCTX-M-15 (13 isolates; 35.1%), followed by blaCTX-M-8 (11 isolates; 29.5%), blaCTX-M-2 (8 isolates; 21.6%), blaCTX-M-14 (3 isolates; 8.1%), and blaCTX-M-9 (2 isolates; 5.4%) as displayed in Table 3. While variants like blaCTX-M-14 andblaCTX-M-9 seemed to be restricted to medical centers located, respectively, in São Paulo and Rio de Janeiro, blaCTX-M-2,blaCTX-M-8, and blaCTX-M-15 were widely distributed. Only four E. coli isolates collected from two medical centers located in São Paulo and another located in Salvador (2 isolates) were shown to carry blaKPC-2. One of these isolates also carried blaCTX-M-8. Among the three P. mirabilis exhibiting the ESBL phenotype, blaCTX-M-2, blaCTX-M-8, and blaCTX-M-15 were identified in three distinct medical centers located in São Paulo and Rio de Janeiro. A single E. cloacae isolate harboring blaKPC-2 was collected in Salvador. In addition to blaKPC-2, this isolate also harbored blaCTX-M-15. No other carbapenemase encoding genes were identified among E cloacae isolates evaluated. In contrast, 11 of 36 (30.6%) E. cloacae isolates were shown to carry blaCTX-M-15.
Distribution of beta-lactamase encoding genes according to bacterial species and medical center location.
| Bacterial Species/β-lactamase encodinggenes (Number) | Number of Isolates detected by geographic location (Number) | Other β-lactamase encoding genes |
|---|---|---|
| E. coli (52) | ||
| ESBL (37) | ||
| blaCTX-M-2 (8) | SP (7), RJ (1) | |
| blaCTX-M-8 (11) | SP (9), RJ (1), Salvador (1) | |
| blaCTX-M-9 (2) | RJ (2) | |
| blaCTX-M-14 (3) | SP (3) | |
| blaCTX-M-15 (13) | SP (11), RJ (2) | |
| Carbapenemase (4) | ||
| blaKPC-2(4) | SP (2), Salvador (2) | |
| E. cloacae (12) | ||
| ESBL (12) | ||
| blaCTX-M-15 (11) | SP (11), RJ (1), Salvador (1) | |
| BlaSHV-12(1) | SP (1) | |
| Carbapenemase (1) | ||
| blaKPC-2(1) | Salvador (1) | |
| P. mirabilis (3) | ||
| ESBL (3) | ||
| blaCTX-M-2 (1) | RJ (1) | |
| blaCTX-M-8 (1) | SP (1) | |
| blaCTX-M-15 (1) | SP (1) | |
| K. pneumoniae (238) | ||
| ESBL (173) | ||
| blaCTX-M-2 (43) | SP (43) | blaKPC-2 (32) |
| blaCTX-M-3 (1) | SP (1) | |
| blaCTX-M-8 (6) | SP (4), RJ (1), Salvador (1) | blaKPC-2 (2) |
| blaCTX-M-9 (3) | SP (3) | |
| blaCTX-M-14 (34) | SP (34) | blaKPC-2 (33) |
| blaCTX-M-15 (84) | SP (56), RJ (17), Salvador (11) | blaKPC-2 (38); blaNDM-1(2) |
| blaCTX-M-35 (2) | SP (2) | |
| blaCTX-M-141 (1) | Salvador (1) | blaKPC-2 (1) |
| AmpC (2) | ||
| blaCMY-2 (1) | SP (1) | |
| blaCMY-141(1) | SP (1) | |
| Carbapenemase (165) | ||
| blaKPC-2 (163) | SP (148), RJ (7), Salvador (8) | |
| blaKPC-3 (1) | SP (1) | |
| blaKPC-30 (1) | SP (1) | |
| blaNDM-1 (2) | RJ (1), Salvador (1) | |
| P. aeruginosa (104) | ||
| PDC (104) | ||
| PDC-1 (4) | SP (2), RJ (1), Salvador (1) | |
| PDC-3 (11) | SP (8), RJ (3) | |
| PDC-5 (15) | SP (13), Salvador (1), RJ (1) | blaIMP-1 in SP (1); blaSPM-1 in SP (2) |
| PDC-6 (1) | SP (1) | |
| PDC-8 (8) | SP (8) | |
| PDC-10 (1) | SP (1) | |
| PDC-11 (3) | SP (1), RJ (1), Salvador (1) | |
| PDC-12 (3) | SP (3) | |
| PDC-16 (11) | SP (9), Salvador (1), RJ (1) | |
| PDC-19A (8) | SP (6), Salvador (2) | blaVIM-2 in SP (1) |
| PDC-24 (2) | SP (2) | |
| PDC-31 (1) | RJ (1) | |
| PDC-35 (19) | SP (19) | blaIMP-74 in SP (1); blaCTX-M-2 in SP (3);blaGES-1 in SP (2); |
| PDC-36 (1) | SP (1) | |
| PDC-37 (4) | SP (3), Salvador (1) | blaKPC-2 in SP (1) |
| PDC-45 (1) | SP (1) | |
| PDC-55 (1) | Salvador (1) | |
| PDC-59 (1) | SP (1) | blaCTX-M-2 in SP (1) |
| PDC-109 (1) | SP (1) | |
| PDC-115 (2) | SP (2) | |
| PDC-120 (1) | SP (1) | |
| PDC-124 (1) | SP (1) | |
| PDC-194 (1) | SP (1) | |
| PDC-218 (1) | SP (1) | |
| PDC-252 (1) | SP (1) | |
| PDC-253 (1) | SP (1) |
SP, São Paulo; RJ, Rio de Janeiro.
Among 238 K. pneumoniae isolates that were eligible for molecular characterization, 165 (69.3%) were shown to carry blaKPC with almost all harboring blaKPC-2. Single isolates carrying blaKPC-3 or blaKPC-30 were collected from a single medical center located in São Paulo. OXA-48, IMP or VIM encoding genes were not detected among the studied isolates. In contrast, two isolates harboring blaNDM-1 were identified in the medical centers located in Salvador and Rio de Janeiro. While ESBL encoding genes, mainly belonging to the CTX-M family, were very frequently detected in K. pneumoniae isolates, plasmid-mediated AmpCs like blaCMY-2 and blaCMY-141 were very rare. The most frequently detected ESBL encoding genes were blaCTX-M-15, blaCTX-M-2, and blaCTX-M-14 as shown in Table 3. In many occasions, blaKPC2 was associated with blaCTX-M-15 (38 isolates), blaCTX-M-14 (33 isolates), and blaCTX-M-2 (32 isolates).
Among the 10 P. aeruginosa isolates exhibiting MICs ≥ 32μg/mL for ceftolozane-tazobactam, five isolates harboring MBL encoding genes were detected with blaSPM-1 (two isolates), blaIMP-1,blaIMP-74, and blaVIM-2 identified, respectively, in single isolates. One P. aeruginosa isolate harboring blaKPC-2 was found and shown to be resistant to ceftolozane-tazobactam, imipenem, and meropenem with MICs of 16, 32 and 16μg/mL, respectively. A high number of Pseudomonas Derived Cephalosporinase (PDC), also denominated AmpC, encoding genes was detected in our collection with PDC-35, -5, -16, -3, being the most frequently observed as shown in Table 3. In contrast, ESBL encoding genes were rarely found in P. aeruginosa, with four isolates from a single center located in São Paulo carrying blaCTX-M-2 and another isolated from a distinct medical center carrying blaGES-1.
The distribution of beta-lactamases according to the participating Brazilian medical center is shown in Fig. 3. The size of each rectangle is proportional to the frequency of each beta-lactamase encoding gene found in the respective medical center. Although K. pneumoniae isolates harboring KPC-2 were found in all medical centers, its frequency varied among the institutions. The same observation can be extrapolated for various PDC types in P. aeruginosa demonstrating the importance of local epidemiology.
DiscussionSurveillance studies are important not only for supporting new drug development but also for determining the antimicrobial susceptibility profile for guiding the selection of the most appropriate antimicrobial therapy, when more comprehensive data are not available (11)]. The enhanced understanding of the resistance mechanisms is a key factor in the search for new antimicrobial agents, allowing for the development of drugs that are effective despite such mechanisms. Ceftolozane-tazobactam is one of the most recently approved cephalosporin-beta-lactamase inhibitor combination for treating infections caused by Gram-negative bacilli, including P. aeruginosa, in Brazil.12 Investigating the susceptibility profile of Brazilian Gram-negative bacilli and the mechanisms of resistance involving these organisms are crucial to support an effective clinical decision.
In our study, ceftolozane-tazobactam showed high antibacterial activity against E. coli with >90% susceptibility regardless of the resistance mechanism. The same favorable resistance profile was observed in E. coli isolated from China,13 Canada14 and US.15 Although this class of drug is not recommended as first line treatment for ESBL infections, the favorable profile renders an alternative for treating ESBL-producing E. coli. Ceftolozane-tazobactam was also very active against P. mirabilis, including the ESBL-producing isolates.15,16 Of note, high clinical cure rates with ceftolozane-tazobactam treatment of IAI and UTI caused by ESBL has been observed in a pooled analysis of ceftolozane-tazobactam clinical trials.17 It is important to notice that ciprofloxacin resistance rates were very high (>50%) among E. coli isolates, emphasizing that this fluoroquinolone should not be prescribed empirically in our setting. Overall, E. cloacae had a low susceptibility rate to ceftolozane-tazobactam, and especially for those isolates resistant to ceftazidime. This result was in concert with the observation of Robin et al.18 The hyperexpression of chromosomal AmpC could justify this result. In addition, association with other mechanisms of beta-lactam resistance could be present in these isolates, such as alteration in outer membrane protein(s), because the susceptibility rate to ertapenem was inferior to those observed for imipenem and meropenem.19 Previous studies have shown that AmpC hyperproducing E. cloacae isolates were usually resistant to ceftolozane-tazobactam.20
Among the five most frequent organisms studied, K. pneumoniae was one of the most difficult to treat pathogen, because these microorganisms were usually resistant to first line treatment drugs like imipenem. The resistance rates to ceftolozane-tazobactam and colistin were also high in our study. This pattern of susceptibility seems to be largely affected by the presence of blaKPC-2, which in many cases was associated with blaCTX-type, ESBL encoding genes belonging to the CTX-M-family. Our resistance rates were higher than those reported previously by Pfaller et al., who had evaluated the activity of cetolozane-tazobactam in bacterial isolates collected in Latin America.21 This difference could be attributed to distinct inclusion criteria like medical centers and the study time period, for example. In fact, it has been widely reported that KPC-2 is endemic in many Brazilian hospitals.4 In our study, blaKPC-2 was the most frequent carbapenemase encoding gene found in the participating medical centers with frequencies varying among the institutions. While blaKPC-3 has been reported frequently in some Latin American countries, it has been found rarely in Brazil.22 In fact, to date, blaKPC-3 has only been reported in A. baumannii isolated from a Brazilian medical center located in São Luís, Maranhão.23 To the best of our knowledge, this is the first study to detect new variants of blaKPC, such as blaKPC-3 and blaKPC-30, in K. pneumoniae in Brazilian medical centers. blaKPC-3 and blaKPC-30 were found to be harbored by K. pneumoniae isolated from urine and intra-abdominal abscess of patients hospitalized at the same Brazilian medical center located in the city of São Paulo. NDM-1-producing K. pneumoniae isolates were unfrequently detected in our study. Only two isolates displaying this genotype were identified: one in urine and another in intra-abdominal abscess of patients hospitalized in Rio de Janeiro and Salvador, respectively.
Ceftolozane-tazobactam has been shown previously to be the most effective agent for P. aeruginosa isolates regardless of resistance to other antimicrobial agents.24 Ceftolozane-tazobactam also demonstrated good activity against isolates non-susceptible to ceftazidime and/or imipenem. The activity of ceftolozane-tazobactam was not affected by hyperproduction of AmpC or other mechanism of resistance as expected.25 In contrast, its activity was affected by production of carbapenemases including metallo-β-lactamases (MBL). In this study, blaKPC-2 was detected in a single P. aeruginosa isolate collected in São Paulo, as observed previously in some medical centers.26–30 In addition, a few MBL-producing P. aeruginosa isolates were detected in our study. SPM-1-producing P. aeruginosa ST277, an MBL producer clone, was widely disseminated in Brazilian hospitals in the 2000s.31 Our results seem to corroborate the results previously reported by Cacci et al., who noticed a decline in the frequency of the SPM-1-producing P. aeruginosa ST277 clone at the intensive care unit of a hospital located in Rio de Janeiro city, where SPM-1 was previously endemic.32 In our study, only two SPM-1-producing P. aeruginosa isolates were identified in two distinct medical centers located in the city of São Paulo. In one of these medical centers, two isolates of P. aeruginosa were shown to harbor genes encoding the IMP variants, blaIMP-1 and blaIMP-74. To the best of our knowledge, blaIMP-74 had not been identified previously in any Brazilian medical center. It was isolated from a urine sample of 44-y-o female patient who had been hospitalized to treat of a urinary tract infection.
ConclusionIn this study, ceftolozane-tazobactam was shown to be very active against E. coli, P. mirabilis and P. aeruginosa isolates and could constitute an excellent therapeutic option including for those isolates resistant to extended-spectrum cephalosporins and carbapenems but not producers of carbapenemases. However, the activity of ceftolozane-tazobactam against K. pneumoniae has been jeopardized by the spread of ESBL and KPC-2-producing K. pneumoniae in Brazilian medical centers. In addition, the co-production of beta-lactamases by such species also compromise the activity of ceftolozane-tazobactam. In this context, surveillance studies like SMART are essential for helping to delineate the changes in the epidemiology of Gram-negative infections over time, not only in Brazil, but also worldwide.
We are very grateful to the Brazilian participating medical centers of the SMART Program for providing bacterial isolates: Adriana Lucia Pires Ferreira; Juvêncio Furtado; Maria Goreth Barberino; Maria Rita Elmor de Araújo; Marinês Dalla Valle Martino; Solange Fujimura.
We thank MSD and IHMA (International Health Management Associates, S.A., Schaumburg, Illinois, U.S.) for providing access to the database of the SMART epidemiological surveillance study.