The Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) pandemic demanded rapid diagnosis to isolate new COVID-19 cases and prevent disease transmission. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) rapidly became the gold standard for diagnosis. However, due to the high cost and delay of the results, other types of diagnosis were implemented, such as COVID-19 Ag Rapid Tests and Reverse Transcription Technique followed by Loop-Mediated isothermal Amplification (RT-LAMP). In this work, we validated the use of RT-LAMP in saliva samples rather than nasopharyngeal swabs, as the collection is more comfortable. First, we selected 5 primer sets based on the limit of detection for SARS-CoV-2 RNA, then validated their sensitivity and specificity in patient samples. A total of 117 samples were analyzed by fluorometric RT-LAMP and compared with qRT-PCR results. Our results show that the use of a high-sensitive primer ORF1-a, together with a low-sensitive primer set Gene E (time to threshold of 22.9 and 36.4 minutes, respectively, using 200 copies of viral RNA), achieved sensitivity in purified RNA from saliva samples of 95.2% (95% CI 76.1‒99.8) with 90.5% specificity (95% CI 69.6‒98.8) (n = 42).As RNA purification increases the turnaround time, we tested the outcome of RT-LAMP utilizing raw saliva samples without purification. The test achieved a sensitivity of 81.8% (95% CI 59.7‒94.8) and a specificity of 90.9% (95% CI 70.8‒98.8). As a result, the accuracy of 92.9% (95% CI 80.5‒98.5) in purified RNA-saliva samples was lowered to an acceptable level of 86.4% (95% CI 72.6‒94.8) in raw saliva. Although mass vaccination has been implemented, new strains and low vaccination progress helped to spread COVID-19. This study shows that it is feasible to track new COVID-19 cases in a large population with the use of raw saliva as sample in RT-LAMP assay which yields accurate results and offers a less invasive test.
Rapid and reliable diagnostic tests are imperative to prevent infectious disease spread and new outbreaks.1 The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has made evident the demand for rapid molecular diagnostics that are universally accessible, scalable, low-cost, and with the proper guarantee of diagnostic accuracy. The molecular diagnosis by Reverse Transcription followed by Polymerase Chain Reaction (RT-PCR) was rapidly implemented for molecular COVID-19 diagnosis and was recommended as the Gold-standard by The World Health Organization (WHO).2,3 However, RT-PCR requires the use of expensive equipment and reagents, along with the need of highly qualified personnel. Moreover, the procedure itself can take several hours to perform, which causes a delay in the release of results. This delay can be further aggravated by the limited availability of materials. An alternative to meet the demand of a rapid test during the COVID-19 pandemic, other molecular tests were developed as the loop-mediated isothermal amplification, LAMP.4,5 This technique is a rapid diagnostic test with high sensitivity that uses a BST DNA polymerase isolated from Geobacillus stearothermophilus to amplify DNA at a constant temperature, between 60°‒65°C. Due to BST high double-strand displacement activity, target DNA amplification can be achieved without thermocycling in up to 30 minutes using up to three pairs of specific primers that recognize between six regions of the target gene.6 As the patent for the use of LAMP [EP 1020534 B] expired in 2019, the technique has become very attractive for the detection of various viral, bacterial, and fungal infections, including SARS-CoV-2.7,8 Novel BST polymerase mutants, that inhibit the activity of the enzyme at room temperature and reduce nonspecific reactions during the test configuration have also potentialized the use of LAMP.9–11
Baek and collaborators[12] developed a LAMP-associated Reverse Transcription Assay (RT-LAMP) to detect SARS-CoV-2.12 With the use of RT-LAMP, it was possible to detect SARS-CoV-2 in patient samples without the need of a separate prior synthesis of cDNA with a detection limit of ∼100 RNA copies, the same limit as for qRT-PCR. Soon this assay was adapted to ease the result in a non-quantitative manner, using colorimetric pH sensitive-dyes, and this was possible due to the high polymerase activity of BST-polymerase that releases pyrophosphate and hydrogen ions readily detectable with different dyes.13 Our goal was to establish a rapid and efficient molecular test for SARS-CoV-2 diagnosis based in the use of saliva as the sample, without the need for saliva pretreatment or RNA purification. The focus was on developing a highly accurate RT-LAMP assay, and the obtained results were compared with those achieved using the gold-standard method of RT-PCR.
Material and methodsClinical samplesNasopharyngeal swabs or saliva were collected from symptomatic individuals evaluated at the COVID-19 Diagnostic Center of the Federal University of Rio de Janeiro (CTD/NEEDIER), between 09/03/2021 and 18/01/2022. All participants were informed about the project and the steps of sample collection and signed written consent. The study was approved by the National Committee of Research Ethics (CAAE 30161620.0.1001.5257). Collection, handling, processing, and disposal of samples followed the biosafety protocols implemented by CTD/NEEDIER ‒ UFRJ. Our cohort comprised 117 individuals, with 39 males and 78 females, with an average age of 39.5 (±15.6) years old. These individuals voluntarily presented with one or several of the following symptoms: fever, cough, dyspnea, sore throat, fatigue, myalgia, joint pain, or gastrointestinal symptoms such as diarrhea. Nasopharyngeal samples from these individuals were previously tested using the lateral flow immunochromatographic platform rapid antigen tests Panbio™ COVID-19 Ag,14 as well as RT-qPCR with N1, N2, and RP primers at the Reference Molecular Virology Laboratory at the Biology Institute, UFRJ. Among these individuals, 74 tested positive and 43 tested negative for SARS-CoV-2 infection between March 2021 and January 2022. Nasopharyngeal swabs for RT-LAMP were collected using two rayon swabs and kept in a 15 mL conical nuclease-free tube, containing 2 mL 0.9% NaCl sterile solution.15 A random saliva collection (1.0‒2.0 mL) was performed in a sterile collection tube, without stimulating saliva production. At least one hour of fasting before the collection was indicated; however, water (200 mL) was provided. Saliva collection took between 3 to 4 minutes. Immediately after collection, samples were placed on ice (4°C) and transported to the laboratory within 2 hours, for direct processing or storage at -80°C, until used. Data of individuals is presented in supplementary material (Table S1).
SARS-COV-2 positive controlsVero cells were infected with SARS-CoV-2 variant RJ1 (GISAID: EPI_ISL_528539) isolated in Rio de Janeiro (Brazil).16–18 Culture-supernatants of infected and uninfected cells were inactivated with 1/100 inactivation solution (0.5 M TCEP, 0.1 M EDTA pH 8.0, and 1.15 N NaOH), followed by boiling at 95°C for 5 min15 and kept frozen at -80°C until used. The copy number was established using as a control a serial dilution of plasmid target provided by IDT containing 200,000 copies/mL of the SARS-Cov2 N gene.14
Sample Inactivation and RNA purificationFrozen samples were thawed and nasopharyngeal swabs (0.14 mL) or saliva (0.6 mL) were transferred to 1.5 mL nuclease-free tubes and buffered with 0.5M TCEP (Tris [2-Carboxyethyl] Phosphine), 0.1 M EDTA Ph 8.0, and 1.15 N NaOH.15 Samples were inactivated at 95°C for 5 minutes, followed by an ice bath for 5 minutes. After inactivation, 140 µL of the sample was purified with QIAamp® Viral RNA kit (QIAGEN, Cat. n° 52906) following the manufacturer´s instructions. Samples were kept under -80°C until used.
RT-LAMP primersSix different primer sets for detecting SARS-CoV-2 by RT-LAMP were chosen from the literature (Table S2). Each set detects different regions of the genome of the SARS-CoV-2 virus, which correspond to the nucleocapsid protein (Gene N and Gene N-A), the viral envelope protein (Gene E), and the ORF1ab polyprotein (Genes ORF1a, ORF1a-HMS and ORF1a-HMSe).15,19–23 These primer sets were designed to detect and amplify regions of viral RNA that are less susceptible to mutation. The set of primers selected for RT-LAMP was purchased from Exxtend Biotecnologia LTDA (Table S2). Primers were resuspended in nuclease-free water to 100 μM and then 150 µL aliquots of each 10 × primer set were prepared as follows: 16 µL FIP, 16 µL BIP, 2 µL F3, 2 µL B3, 4 µL LF and 4 µL LB.
RT-LAMP reactionsRT-LAMP reactions were performed following New England Biolabs’ recommended protocol using: 6.25 µL WarmStart Colorimetric® LAMP 2 × Master Mix (DNA & RNA) (M1800) or WarmStart® LAMP Kit (DNA & RNA) (E1700), combined with 1.25 µL 10 µM SYTO™ 9 Green Fluorescent Nucleic Acid Stain (Invitrogen™ #S34854), 1.25 µL 10 × primer mix, and 2.75 µL DEPC-treated water. Reactions were measured in StepOne Plus™ Real-Time PCR System (Applied Biosystems™ #4376600) at 65°C. An amplification signal was acquired every 15 seconds for 240 “cycles” with a total incubation time of around 86 minutes. Samples were analyzed in duplicate in the same microplate. Otherwise noted, 200 copies of SARS-CoV-2 purified RNA were used as a positive control. Water nuclease-free was used as a negative control, at least in triplicate, at different positions in the microplate. Data were acquired using the StepOne™ Plus v2.3 software and analyzed with GraphPad Prism (version 8.1.1; GraphPad Software). Time to Threshold (Tt) was determined from the real-time fluorescence signal with a cutoff of 40 minutes.
Primer alignment with SARS-CoV-2 omicron variantsThe SARS-CoV-2 isolates within Brazil, deposited at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genome/viruses/variation/30May2023), were analyzed. The selected Omicron sub-variants isolated in 2023 with a largest number of deposited sequences were XBB.1.9 and XBB.1.5. The region between the Forward and Backward outer primers (F3-B3) from each primer set was retrieved and aligned to the NCBI reference sequence GCF_009858895.2 (NC_045512.2) (SARS-CoV-2 isolate Wuhan-Hu-1). The Single Nucleotide Polymorphisms (SNPs) were then aligned to the individual primers of each set using the NCBI BLASTn suite.24 The nucleotide alignment was built at NCBI database while the phylogenetic tree was built using iTol tool.25
Data and statistical analysisAll data were analyzed with GraphPad Prism (version 8.1.1; GraphPad Software). Sensitivity, specificity and positive predictive values, and negative predictive values were calculated as in Lalkhen et al. 2008.26 The 95% Confidence Interval was calculated using the confidence interval for a proportion calculator.27
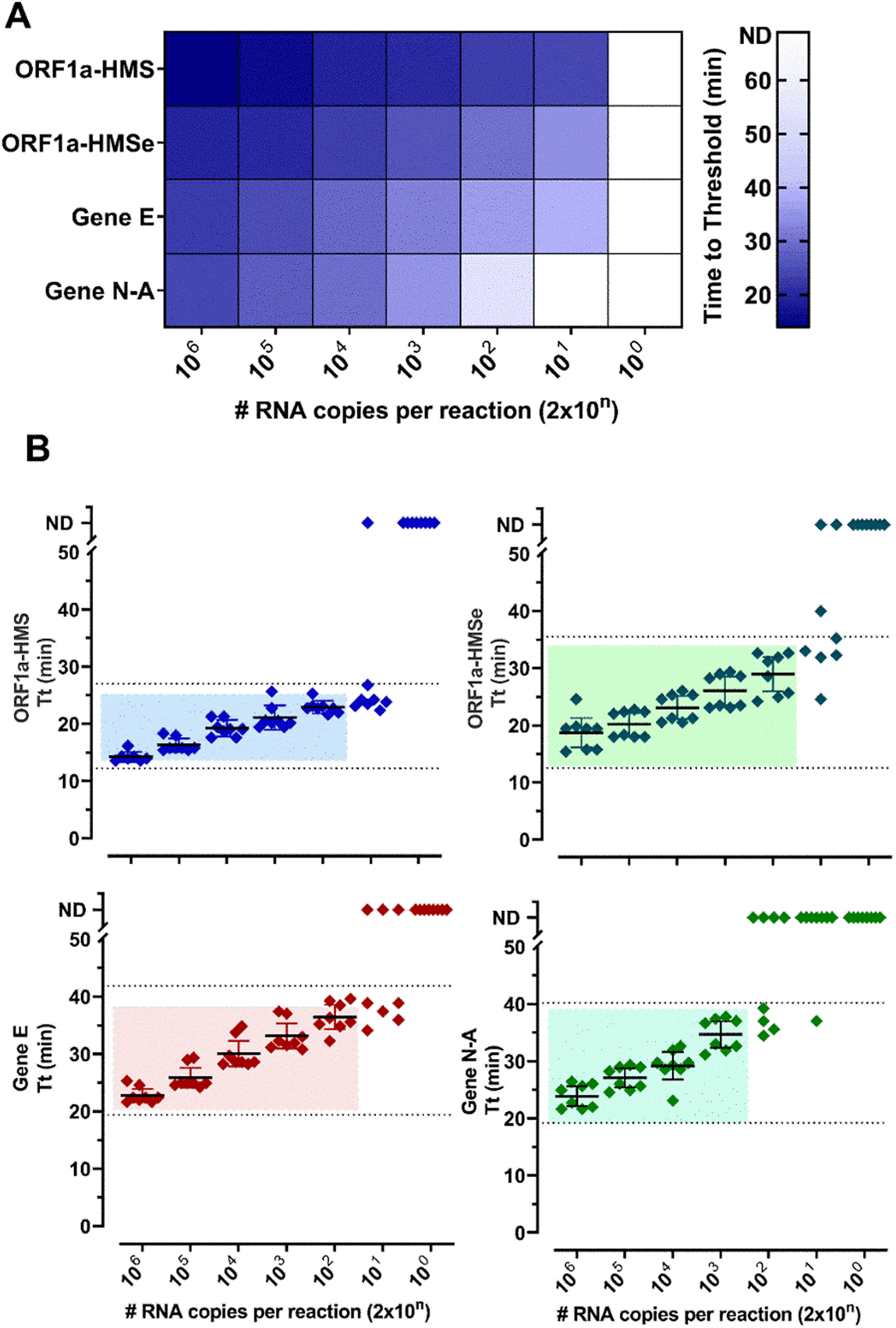
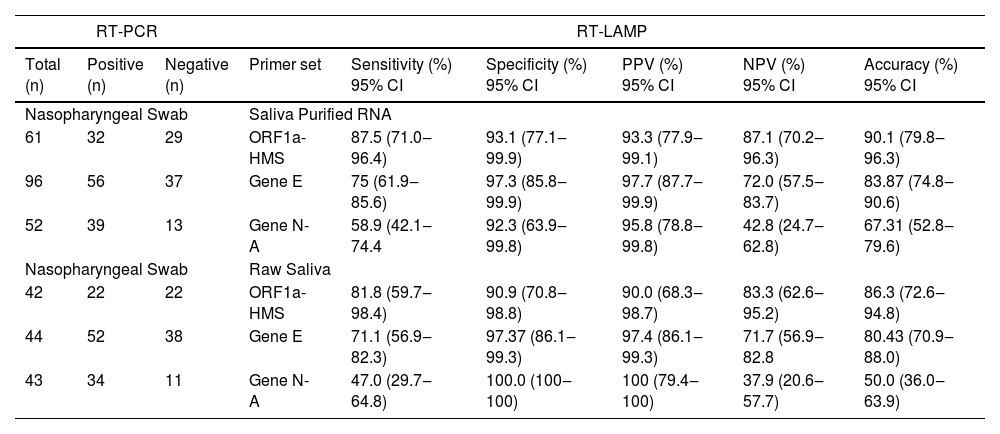
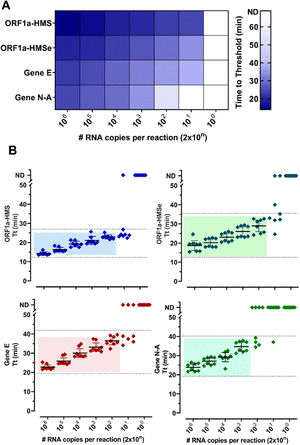
ResultsSelection of primer sets and comparison of RT-LAMP assay using nasopharyngeal swabs or salivaThe Limit of Detection (LoD) of six primer sets was determined (Table S2) and (Fig. 1). The primer sets, that detected at least 200 copies of viral RNA in less than 40 minutes, are shown in Fig. 1. The results indicate that each primer set has a different LoD and a low rate of false positives, except for Gene N, which gave positive results in non-RNA controls and Gene ORF1a, which did not present positive amplification. We discontinued the use of these two primer sets. To detect SARS-CoV-2 in patient samples we chose to use primer sets ORF1a-HMS, ORF1a-HMSe, and Gene E, which had different sensitivities with a time to Threshold (Tt), to detect 200 copies of 22.9 ± 1.1; 29.0 ± 3.6; 36.4 ± 2,5 minutes, respectively, and Gene N-A, which had Tt of 34.7 ± 2.8 minutes to detect 200 copies. Although Gene N-A had a lower detection limit, we chose to use this primer as in our experience, highly sensitive primers are more prone to nonspecific amplification.
Primer performance for the detection of SARS-CoV-2 by RT-LAMP: Increasing concentrations of SARS-CoV-2 titered samples were subjected to RT-LAMP using primer sets ORF1a-HMS, ORF1a-HMSe, Gene E and Gene N-A. (A) Heat-map representation of time to threshold values and (B) Time to threshold values of eight replicates for each oligonucleotide set performed in three independent experiments. Water was used as negative control (data not shown), and in reactions where no amplification was recorded, the “Time to Threshold” is reported as Not Detected (ND), Colored areas highlight the time to the threshold and #RNA copies with 100% positive detection.
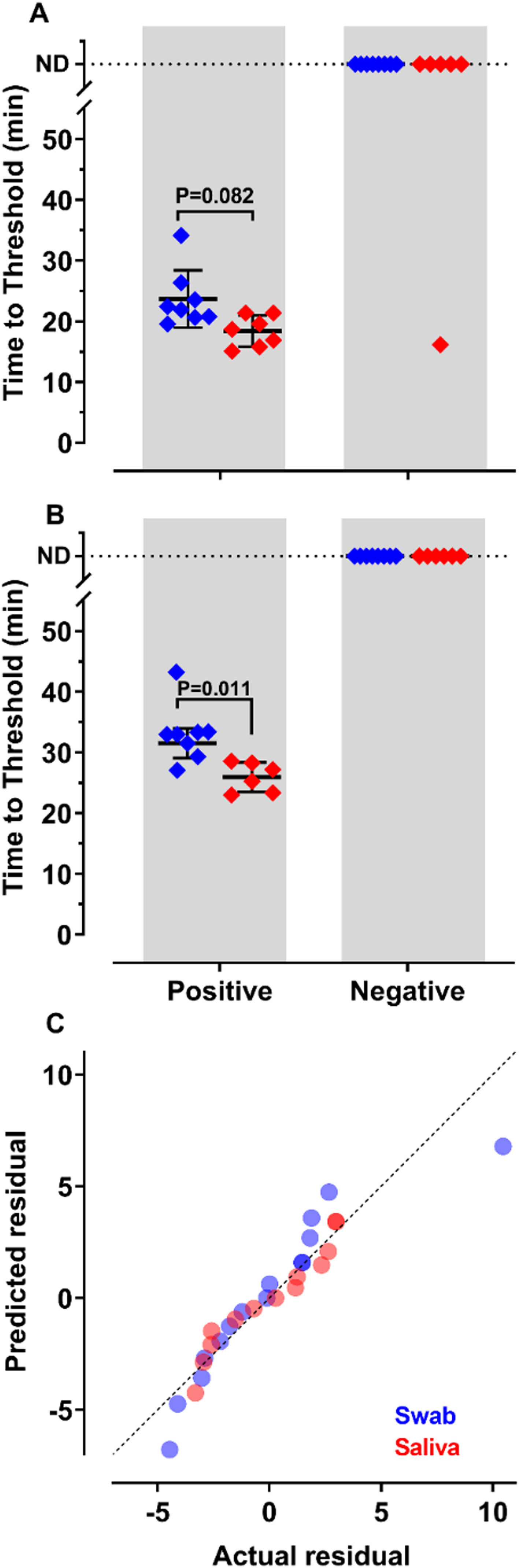
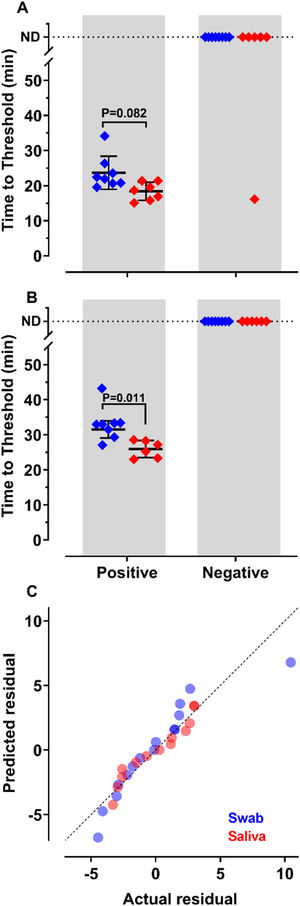
To decide if saliva samples were amenable to RT-LAMP in our settings, we compared the detection of SARS-CoV-2 in these samples with those prepared from nasopharyngeal swabs. Results show that sensitivity and accuracy of detection were almost indistinguishable among the two protocols (Fig. 2A and B). The Q-Q plots (Fig. 2C) show that the distribution is similar between both samples. Nevertheless, the test performed with saliva had faster Tt (p = 0.011), using primer set Gene E. The variances of the results were significantly higher for samples prepared from nasopharyngeal swabs than for those obtained from saliva (Fig. 2C). These results indicate that it is possible to create a saliva-based diagnostic test kit that can be applied without the discomfort caused by nasopharyngeal swabs.
of SARS-CoV-2 from nasopharyngeal swabs or saliva samples by RT-LAMP: RNA was purified from nasopharyngeal swabs (blue) or saliva (red) from both qPCR positive (n = 12) and negative (n = 16) SARS-CoV-2 samples. RT-LAMP assay was performed with primers set ORF1a-HMSe (A) and Gene E (B) at 65°C. Reactions were performed at least in triplicate in two independent experiments. Water was used as a negative control and 200 viral copies per reaction were used as a positive control (data not shown). In reactions where no amplification was recorded, the “Time to Threshold” is reported as Not Detected (ND). The black lines represent the Mean and standard deviation (*p = 0.001). (C) Distribution profile of purified RNA from nasopharyngeal swab samples (blue) and purified RNA from saliva (red) between actual versus predicted residual in the Q-Q plot.
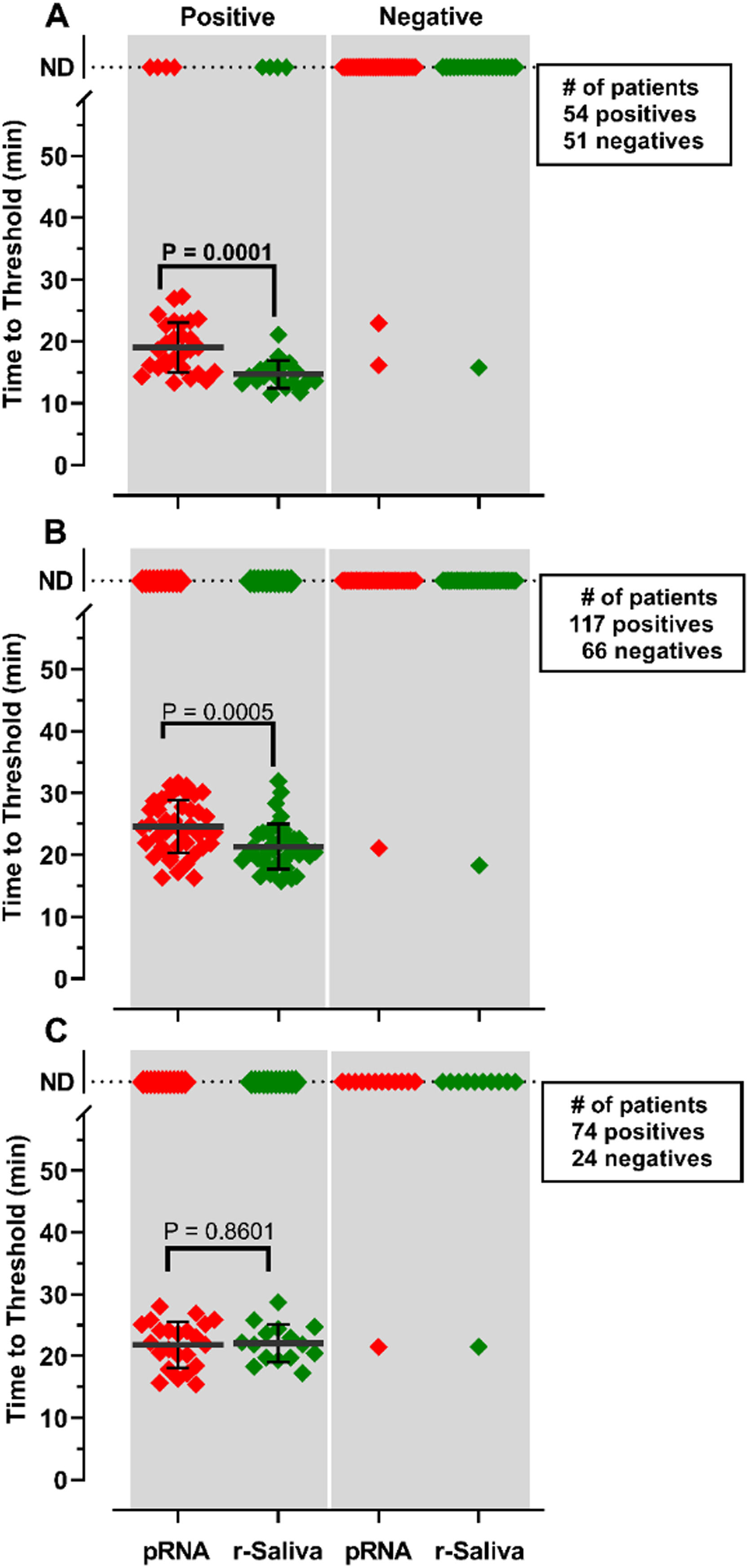
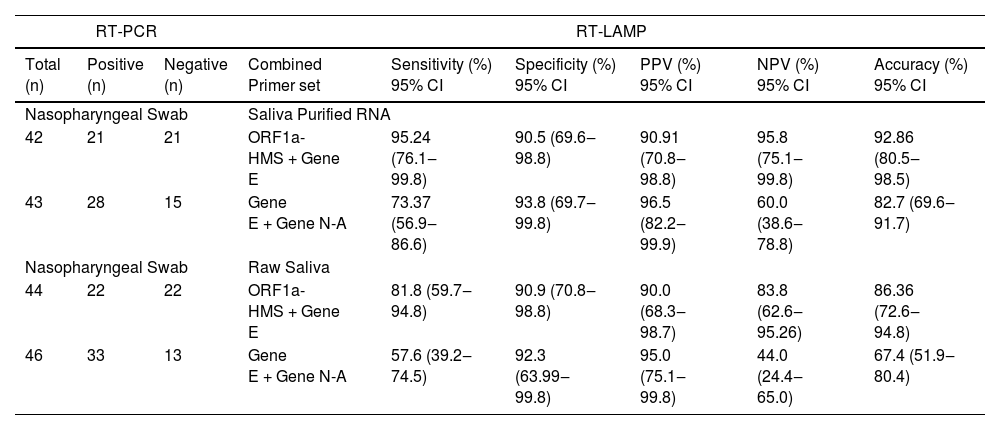
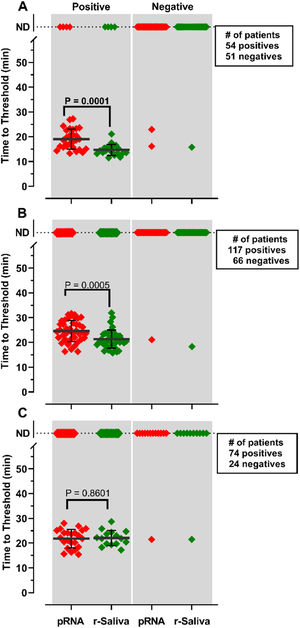
RNA purification from nasopharyngeal samples from COVID-19 patients, is performed in the gold-standard qRT-PCR assay. However, RNA purification increases the risk of human contamination and is time-demanding unless automated handling is used. Therefore, we compared the RT-LAMP assay between purified saliva-RNA (pRNA) and raw saliva (r-saliva) samples (Fig. 3). The time to threshold value was significantly lower in the former with a difference of approximately 3 minutes, when assayed with ORF1a-HMS and Gene E sets (Fig. 3A and B), while no difference was observed with Gene N-A set (Fig. 3C). It can also be observed that false negatives were not different between the two samples. The sensitivity, specificity, positive/negative predictive values, and accuracy were tested independently for each primer set (Table 1). The primer set that outperformed the best was ORF1a-HMS, in both samples. The accuracy with saliva-purified RNA was 90.1% (95% CI 79.8‒96.3) compared to raw saliva which was 86.3% (95% CI 72.6‒94.8). Gene E performance was within the acceptable range between 83.9% (95% CI 74.8‒90.6) and 80.4% (95% CI 70.9‒88.0) in saliva purified RNA and raw saliva, respectively. As expected, Gene N-A sensitivity was low, accordingly to the calculated LoD value, with an accuracy lower than 70% in both samples.
Comparison of detection of SARS-CoV-2 in RNA purified from saliva samples vs raw saliva by RT-LAMP: SARS-CoV-2 was detected in RNA purified from saliva samples (pRNA; red) or raw saliva (r-saliva; green) from both qPCR positive or negative SARS-CoV-2 samples by RT-LAMP with the primer sets (A) ORF1a-HMSe, (B) Gene E or (C) Gene N-A. Water was used as a negative control and 200 viral copies per reaction were used as a positive control (data not shown). In reactions where no amplification was recorded, the “Time to Threshold” is reported as Not Detected (ND). The black lines represent the mean and standard deviation. Reactions were performed at least in triplicate in two independent experiments.
Comparison of RT-LAMP assay in saliva purified RNA vs. raw saliva diagnosed with an individual primer set.
After analyzing the results with individual primers, the pairwise statistics combining the results of two different primers were calculated. For this purpose, we performed a pairwise assay with the two more sensitive primer sets ORF1a-HMS/Gene E or the two less sensitive primer sets Gene E/Gene N-A (Table 2). We considered the result positive if either one or both primer sets were positive in the same sample, and negative if no detection was observed in the same sample with both primer sets. The sensitivity of ORF1a-HMS/Gene E performed better with saliva-purified RNA samples than with raw saliva, with a sensitivity of 95.2% (95% CI 76.1‒99.8) compared to 81.8% (95% CI 59.7‒94.8). It was also outperformed when compared to the Gene E/Gene N-A pair, which attained a sensitivity lower than 85% in either sample. Comparison of the two primer sets ORF1a-HMS/Gene E with individual ORF1a-HMS indicates that the use of the two primers is the best protocol for the diagnosis of SARS-CoV-2 as it increases the sensitivity of RT-LAMP, from 87.5% to 95.2%, for the reactions using purified RNA from saliva.
Comparison of RT-LAMP assay in saliva purified RNA vs. Raw saliva diagnosed with a pair set of primers.
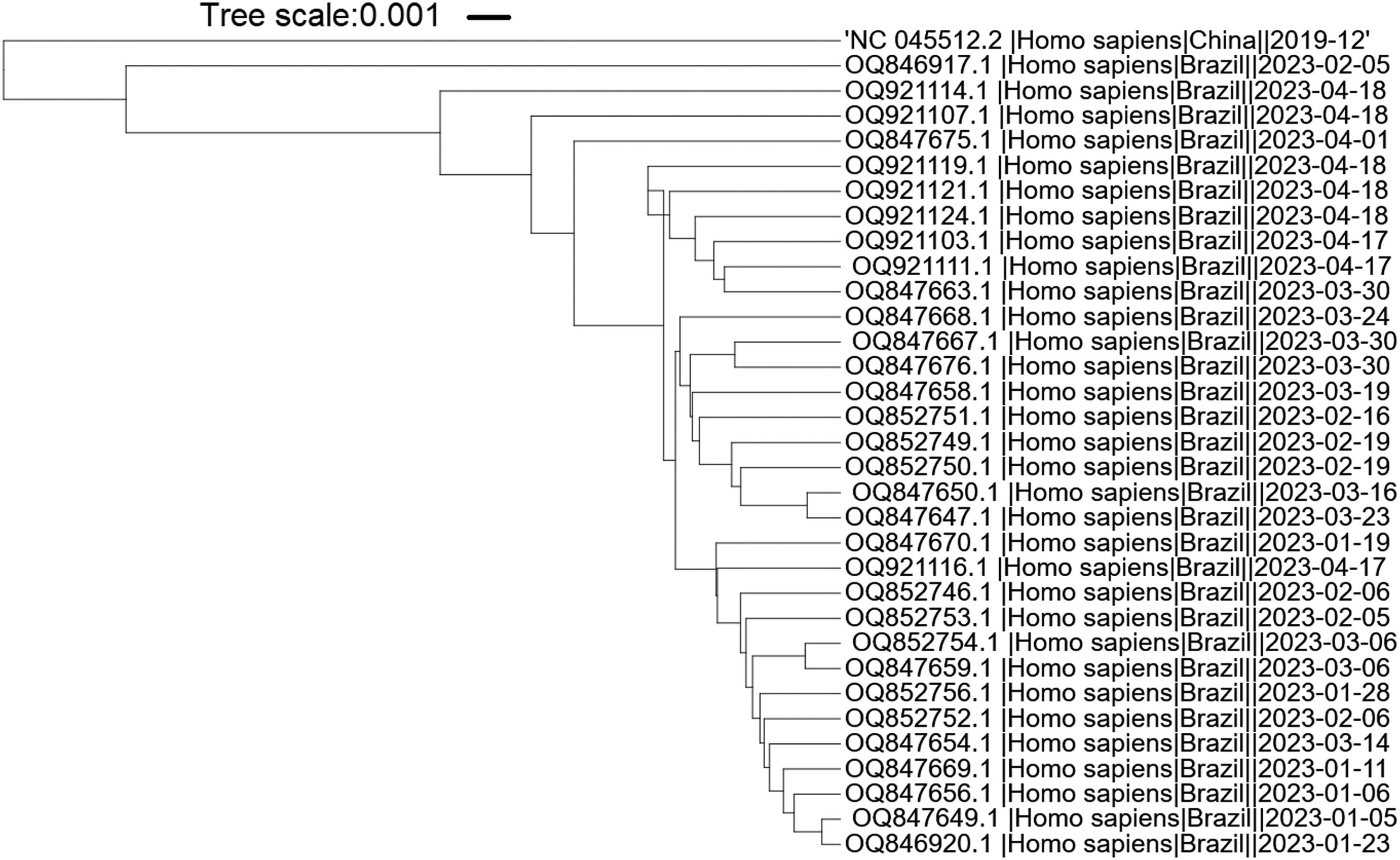
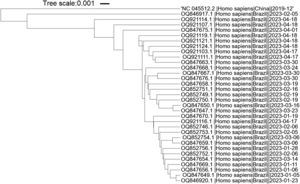
It is important to highlight that SARS-CoV-2 has undergone evolutionary changes since its initial emergence, resulting in the identification of new variants and subvariants worldwide. In order to assess the suitability of the primers utilized in this study for the new Omicron variants, we specifically selected the predominant Brazilian subvariants XBB.1.5 and XBB.1.9 isolated during the first five months of 2023. The phylogenetic tree shows that the variants and reference sequence form to clusters (branch length of 0.033), while among the Omicron variants themselves, there are minimal changes (branch length of 0.0085) (Fig. 4). The sequence region between the F3 and B3 primers of each primer set was aligned against these subvariants. A total of nineteen sequences related to XBB.1.9 and fifteen sequences related to XBB.1.5 were subjected to analysis. Interestingly, only two sequences belonging to XBB.1.9 (accession numbers OQ847669 and OQ847670) and one sequence belonging to XBB.1.5 (accession number OQ847676) exhibited a single SNP in the ORF1a-HMS-LF primer and the B3 primer, respectively. Furthermore, all subvariants displayed a single SNP in the Gene E-FIP (F1c) region, while no SNPs were observed in the Gene N-A primer region.
DiscussionThe RT-LAMP technique is a novel assay that has been proposed for the diagnoses of viral diseases such as dengue and zika7,10,28 but it was studied thoroughly during the SARS-CoV-2 pandemic. The technique rapidly evolved to be used as a point-of-care test, with a colorimetric readout.19,13,29 This feature was novel as it does not depend on fluorometric readers as qRT-PCR. New England Biolabs (NEB) released a kit that exhibited detection over 30 minutes combined with colorimetric visualization, the WarmStart® Colorimetric LAMP 2 × Master Mix kit (DNA & RNA, M1800 ‒ NEB). However, although using a colorimetric method lowers costs related to equipment, it relies on low-buffered media during testing that can increase RNA degradation and decrease the activity of BST-polymerase. It also relies on visual detection, which can lead to false results. RT-LAMP for COVID-19 diagnosis was also improved using more amenable samples, such as saliva. Rabe and Cepko,15 and Yu et al.30 used saliva samples with a previous RNA extraction, while Lalli et al.22 and Kobayashi et al.31 reported an assay preserving RNA by the addition of proteinase K or guanidine/DTT to saliva samples, respectively. Several modifications to this test have been discussed. Thi et al.5 suggest that the RT-LAMP assay, when combined with LAMP sequencing, is suitable for the analysis of large numbers of RT-LAMP reactions due to the fully scalable DNA barcoding strategy. As more sensitive primer sets become available, the application of the RT-LAMP assay has great potential.
To set up the RT-LAMP technique in our setting, we used the N primer set in our first assays (Table 1).19 This particular primer set initially showed promising colorimetric readouts and several specific DNA products that were analyzed by agarose gel electrophoresis (data not shown). However, after multiple experiments, we encountered false positive results, primarily attributed to contamination resulting from aerosols formed during manipulation of agarose gels. To address this issue and obtain more accurate results, we made the decision to adopt fluorometric readouts as an alternative to colorimetric and agarose gels readings. Fluorometric readouts offer the advantage of enabling semi-quantitative analysis of the data, allowing us to work with time threshold results instead of relying solely on visual readings. This approach minimizes the risk of contamination and provides a more precise and reliable assessment of the amplification results.
Different sets of specific primers have been used for COVID-19 diagnosis, which ensure that the designed primers have high specificity to the target sequence, do not have significant complementarity to other non-target sequences in the sample and that do not match to regions with high variability among different isolates, however the limit of detection between reported primers was not tested. We tested the LoD of four sets of primers already reported in the literature, which target different viral regions (Table 1, Fig. 1). Evaluation confirms that these primers can amplify as less as 200 viral copies in up to 40 mins at 65°C as reported,6,8,15,21,29 however the specificity of the primers towards SARS-CoV-2 was different between them. Regarding this set of primers, it should be noted that at the course of experiments, the ORF1a-HMS primer set was also prone to false-positive results in some laboratory settings. Having said that, we recommend the use of primers with high specificity with care to avoid contamination. We also recommend the design of more sets of primers for RT-LAMP diagnosis, which can readily substitute those that give false positive results.
After comparing the LoD of the different primer sets, we selected to use RT-LAMP fluorometric readout (Syto9™) and to test each sample with at least two primer sets. This experimental protocol was used to compare the accuracy by using RNA isolated from saliva samples and nasopharyngeal swabs, obtained from patients investigated for COVID-19. Fig. 2 shows that purified RNA samples from nasopharyngeal swabs or saliva correlates very well with those reported by RT-PCR. Although the RT-LAMP Tt values were statistically different between both samples, they confirm that saliva can be used in the diagnosis of COVID-19 in place of nasopharyngeal swabs.22,32,33 Overall results show that the use of the two primer sets (ORF1a-HMS/Gene E) performed better in purified RNA from saliva than in raw saliva samples. However, the latter attained an acceptable range with a sensitivity and specificity of 81.8% (95% CI 59.7‒94.8) and 90.9% (95% CI 70.8‒98.8), respectively. Amid the SARS-CoV-2 pandemic, numerous molecular tests based on RT-LAMP were developed. However, each protocol had to be individualized for each laboratory setting. We show that the specificity of our test is comparable with RT-LAMP results that used nasopharyngeal swabs with a specificity of 92% (95% CI 85‒96).34 These results showed that the accuracy of RT-LAMP using purified RNA or raw saliva was like that of RT-PCR but required less time under isothermal conditions. Although the use of RT-LAMP to detect molecular components of the virus in raw saliva is less sensitive it can be useful for rapid diagnosis because viral load is reported to be still high within 7 days after the onset of SARS-CoV-2 infection. Our results corroborate those of other authors who also detected the presence of viral RNA copies with appropriate sensitivity using the RT-LAMP assay. Concerning the choice between RT-LAMP or rapid tests, such as the Panbio™ COVID-19 Ag test that has been widely used in Brazil, both can be comparable as the overall reported sensitivity and specificity for the PanbioTM test was 84.3% (95% CI 75‒93.8) and 98.2% (95% CI 96‒98.8), respectively.35 As the accuracy between RT-LAMP and rapid tests is comparable, the choice depends on the specific requirements. RT-LAMP may be preferred in settings where high sensitivity and specificity are crucial, and where RT-PCR is costly. Rapid tests, on the other hand, may be more suitable when immediate results are needed, however concomitant assays are desirable.
It is important to note that SARS-CoV-2 has undergone several evolutionary changes since its initial emergence, resulting in the isolation of various new variants worldwide. The WHO Virus Evolution Working Group has classified these variants into two categories: Variants of Interest (VOIs) and Variants of Concern (VOCs). In Brazil, from March 2020 to May 2023, a total of 6,584 nucleic acid sequences isolated from nasopharyngeal sample have been deposited at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genome/viruses/variation/) from the VOC group. Since both RT-PCR and RT-LAMP assays rely on highly specific primers for the diagnosis of COVID-19, it is crucial to review the primer sets used to account for new variants. The primer sets employed in this study are highly specific for the reference isolate Wuhan-Hu-1. However, it is worth noting that mismatches within the primers, particularly near the 3′ end, can lead to a complete failure of LAMP amplification of the target sequence. Analysis of the new Omicron variants isolated in Brazil in 2023 revealed that some primers had a Single Nucleotide Polymorphism (SNP) compared to these variants. Although the mismatches were not located at the 5′ or 3′ end of the primers, they can still potentially affect the sensitivity of the RT-LAMP assay. A recent publication has described a modification of the RT-LAMP assay, which incorporates a minimal amount of high-fidelity DNA polymerase with 3´exonuclease activity into the reaction system.35 This modification allows for the tolerance of nucleotide mismatches at the 3´ end of primers and has the potential to be utilized in the diagnosis of new SARS-CoV-2 variants. Therefore, it is crucial to monitor the evolution and emergence of new SARS-CoV-2 variants to design new primers that exhibit a high degree of complementarity to the target sequence.
In conclusion, our study shows that the RT-LAMP assay for SARS-CoV-2 diagnosis can be set up readily using raw saliva samples. We recommend the use of at least two sets of primers, with different efficiencies for each sample and surveillance of new variants should always be considered.
We would like to thank Professor André Felipe Andrade dos Santos (Department of Genetics, Institute of Biology, UFRJ) for providing the NB-2 laboratory; the working group of Centro de Triagem e Diagnóstico (CTD/NEEDIER ‒ UFRJ) for collecting patient samples; Rafael Alves Andrade and Jessica Moreira for the technical support; the Synthetic Biology team Osiris-Rio ‒ UFRJ and Prof. Luis Maurício Trambaioli da Rocha e Lima for helpful discussions. This work was supported by grants from Emenda Parlamentar n° 40540013 and 40540021 to MML and FAC; Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) CHAMADA C ‒ AÇÃO EMERGENCIAL COVID-19 to MSA, Pos-doc Fellowship FAPERJ E26/ 210.195/2020 to KMS, Cientistas do Nosso Estado CNE FAPERJ E26/201.070/2021 to MML and Programa de Desenvolvimento Tecnológico em Medicina de Precisão FAPERJ E-26/210.658/2021 to TMPPC.